Alternative Lengthening of Telomeres
Alternative Lengthening of Telomeres (also known as "ALT") is a telomerase-independent mechanism by which cancer cells avoid the degradation of telomeres.
Background
At each end of the chromosomes of most eukaryotic cells, there is a telomere: a region of repetitive nucleotide sequences which protects the end of the chromosome from deterioration or from fusion with neighboring chromosomes. At each cell division, the telomeres get shorter, eventually preventing further cell division. Healthy adult somatic cells in mammals do not have active telomerase enzymes, so that cancer cells stop proliferating unless they have a mutation which restores the telomeres. Often, this is due to a telomerase enyme being reactivated, but alternative mechanisms also occur.
Mechanism of recombination-mediated telomere synthesis
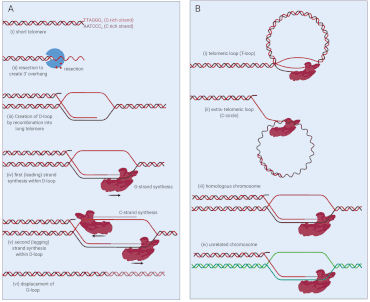
The main alternative lengthening mechanism for telomeres is a type of homologous recombination called Break-induced Telomere Synthesis (or BITS).[1] Normally, homologous recombination allows broken DNA strands to be repaired by lining up with a matching sequence of undamaged DNA, but in BITS, this mechanism is used to extend telomeres. Because telomeres are by nature repetitive, matching sequences are widely available.
In proposed models for how BITS works, the process begins with the resection of a damaged telomere end: one of the strands is cut away to provide a single strand of DNA (the Guanosine-rich strand) that can bind to into a matching (homologous) template, forming a so-called displacement loop (D-loop) (Figure 1a).[2] In ALT, there is evidence that this template consists of: (i) a centromere proximal sequence of the same chromosome (T-loop), (ii) circular extrachromosomal telomeric sequences (C-circles), (iii) homologous chromosomes, or (iv) other chromosomes (Figure 1b). ALT may arise from a combination of some or all of these templates.[3] Importantly, because telomeres are highly repetitive, invasion between or within telomeres is not limited by the requirement for extended homology in homologous recombination. After D-loop formation, DNA polymerase δ extends the invaded G-strand end, copying material beyond the original breakpoint, leading to initiation of lagging strand synthesis of the C-strand, also by DNA polymerase δ.[4]
The second feature of ALT is the production of a non-conservative DNA product at the telomere. At the conclusion of the copying reaction, both strands contain entirely new DNA. This is different from normal ‘semi-conservative’ DNA replication, where one strand is newly synthesized, and the other comes from the original template. In this manner, ALT allows entire telomeric sequences to be copied from one chromosome to another, without affecting the length or integrity of the copied sequence. Recent work suggests that ALT DNA copying (BITS) proceeds via a D-loop migration model, which is supported by the observation of non-conservative rather than semi-conservative products of break-induced replication at ALT telomeres[5] and the D-loop-shaped products observed in two-dimensional gel electrophoresis at sites undergoing BIR.[6]
References
- O'Rourke, JJ; Bythell-Douglas, R; Dunn, EA; Deans, AJ (December 2019). "ALT control, delete: FANCM as an anti-cancer target in Alternative Lengthening of Telomeres". Nucleus (Austin, Tex.). 10 (1): 221–230. doi:10.1080/19491034.2019.1685246. PMC 6949022. PMID 31663812.
 Text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License. - Zhang, JM; Yadav, T; Ouyang, J; Lan, L; Zou, L (22 January 2019). "Alternative Lengthening of Telomeres through Two Distinct Break-Induced Replication Pathways". Cell Reports. 26 (4): 955–968.e3. doi:10.1016/j.celrep.2018.12.102. PMC 6366628. PMID 30673617.
- O’Rourke, Julienne J; Bythell-Douglas, Rohan; Dunn, Elyse A; Deans, Andrew J (1 January 2019). "ALT control, delete: FANCM as an anti-cancer target in Alternative Lengthening of Telomeres". Nucleus. 10 (1): 221–230. doi:10.1080/19491034.2019.1685246. PMC 6949022. PMID 31663812.
- Donnianni, RA; Zhou, ZX; Lujan, SA; Al-Zain, A; Garcia, V; Glancy, E; Burkholder, AB; Kunkel, TA; Symington, LS (7 November 2019). "DNA Polymerase Delta Synthesizes Both Strands during Break-Induced Replication". Molecular Cell. 76 (3): 371–381.e4. doi:10.1016/j.molcel.2019.07.033. PMC 6862718. PMID 31495565.
- Min, J; Wright, WE; Shay, JW (15 October 2017). "Alternative Lengthening of Telomeres Mediated by Mitotic DNA Synthesis Engages Break-Induced Replication Processes". Molecular and Cellular Biology. 37 (20). doi:10.1128/MCB.00226-17. PMC 5615184. PMID 28760773.
- Sneeden, JL; Grossi, SM; Tappin, I; Hurwitz, J; Heyer, WD (May 2013). "Reconstitution of recombination-associated DNA synthesis with human proteins". Nucleic Acids Research. 41 (9): 4913–25. doi:10.1093/nar/gkt192. PMC 3643601. PMID 23535143.