Aspergillus sclerotioniger
Aspergillus sclerotioniger is a species of fungus in the genus Aspergillus. It belongs to the group of black Aspergilli which are important industrial workhorses.[2][3][4] A. sclerotioniger belongs to the Nigri section. The species was first described in 2004.[1] It has been found in green coffee beans from India. It is a very effective producer of ochratoxin A and ochratoxin B, and produces aurasperone B, pyranonigrin A, corymbiferan lactone-like exometabolites, and some cytochalasins. The genome of A. sclerotioniger was sequenced and published in 2014 as part of the Aspergillus whole-genome sequencing project – a project dedicated to performing whole-genome sequencing of all members of the genus Aspergillus.[5] The genome assembly size was 36.72 Mbp.[5]
| Aspergillus sclerotioniger | |
|---|---|
| Scientific classification | |
| Kingdom: | Fungi |
| Division: | Ascomycota |
| Class: | Eurotiomycetes |
| Order: | Eurotiales |
| Family: | Trichocomaceae |
| Genus: | Aspergillus |
| Species: | A. sclerotioniger |
| Binomial name | |
| Aspergillus sclerotioniger Samson & Frisvad (2004)[1] | |
Growth and morphology
Aspergillus sclerotioniger has been cultivated on both Czapek yeast extract agar (CYA) plates and Malt Extract Agar Oxoid® (MEAOX) plates. The growth morphology of the colonies can be seen in the pictures below.
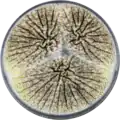 Aspergillus sclerotioniger growing on CYA plate
Aspergillus sclerotioniger growing on CYA plate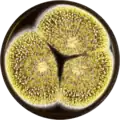 Aspergillus sclerotioniger growing on MEAOX plate
Aspergillus sclerotioniger growing on MEAOX plate
References
- Samson, R. A. New ochratoxin A or sclerotium producing species in Aspergillus section Nigri. Studies in Mycology 2004, 50, 45–61.
- Pel, H. J.; de Winde, J. H.; Archer, D. B.; Dyer, P. S.; Hofmann, G.; Schaap, P. J.; Turner, G.; de Vries, R. P.; Albang, R.; Albermann, K.; Andersen, M. R.; Bendtsen, J. D.; Benen, J. A. E.; van den Berg, M.; Breestraat, S.; Caddick, M. X.; Contreras, R.; Cornell, M.; Coutinho, P. M.; Danchin, E. G. J.; Debets, A. J. M.; Dekker, P.; van Dijck, P. W. M.; van Dijk, A.; Dijkhuizen, L.; Driessen, A. J. M.; d’Enfert, C.; Geysens, S.; Goosen, C.; Groot, G. S. P.; de Groot, P. W. J.; Guillemette, T.; Henrissat, B.; Herweijer, M.; van den Hombergh, J. P. T. W.; van den Hondel, C. A. M. J. J.; van der Heijden, R. T. J. M.; van der Kaaij, R. M.; Klis, F. M.; Kools, H. J.; Kubicek, C. P.; van Kuyk, P. A.; Lauber, J.; Lu, X.; van der Maarel, M. J. E. C.; Meulenberg, R.; Menke, H.; Mortimer, M. A.; Nielsen, J.; Oliver, S. G.; Olsthoorn, M.; Pal, K.; van Peij, N. N. M. E.; Ram, A. F. J.; Rinas, U.; Roubos, J. A.; Sagt, C. M. J.; Schmoll, M.; Sun, J.; Ussery, D.; Varga, J.; Vervecken, W.; van de Vondervoort, P. J. J.; Wedler, H.; Wösten, H. A. B.; Zeng, A.-P.; van Ooyen, A. J. J.; Visser, J.; Stam, H. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat. Biotechnol. 2007, 25, 221–231, doi:10.1038/nbt1282.
- Goldberg, I.; Rokem, J. S.; Pines, O. Organic acids: old metabolites, new themes. Journal of Chemical Technology & Biotechnology 2006, 81, 1601–1611, doi:10.1002/jctb.1590.
- Pariza, M. W.; Foster, E. M. Determining the Safety of Enzymes Used in Food Processing. Journal of Food Protection 1983, 46, 453–468, doi:10.4315/0362-028X-46.5.453.
- https://genome.jgi.doe.gov/Aspscl1/Aspscl1.home.html