David M. Sabatini
David M. Sabatini is an American scientist and Professor of Biology at the Massachusetts Institute of Technology as well as a member of the Whitehead Institute for Biomedical Research. He has been an investigator of the Howard Hughes Medical Institute since 2008 and was elected to the National Academy of Sciences in 2016. He is known for his important contributions in the areas of cell signaling and cancer metabolism, most notably the discovery and study of mTOR, a protein kinase that is an important regulator of cell and organismal growth that is deregulated in cancer, diabetes, as well as the aging process.
David M. Sabatini | |
|---|---|
| Born | January 27, 1968 |
| Nationality | American |
| Citizenship | United States |
| Alma mater | Brown University Johns Hopkins School of Medicine |
| Known for | Discovery and study of mTOR |
| Scientific career | |
| Fields | Biochemistry Cell Biology |
| Institutions | Whitehead Institute Massachusetts Institute of Technology Broad Institute |
| Doctoral advisor | Solomon Snyder |
Biography
David M. Sabatini was born and raised in New York to Dr. David D. Sabatini and Dr. Zulema Sabatini, both Argentine immigrants from Buenos Aires. He obtained his B.S. from Brown University followed by both his MD and his Ph.D. at Johns Hopkins School of Medicine in Baltimore, Maryland, where he worked in the lab of Solomon H. Snyder. He joined the Whitehead Institute as a Whitehead Fellow in 1997, the same year he matriculated from Johns Hopkins.[1] In 2002 he became an Assistant Professor at MIT and a Member of the Whitehead Institute. He was promoted to tenured professor in 2006.
Sabatini currently resides in Cambridge, Massachusetts and is an avid biker and gardener. His father, David D. Sabatini, is a cell biologist and Professor at New York University. His younger brother, Bernardo L. Sabatini, is a neuroscientist and Professor at Harvard Medical School.[1][2]
Sabatini is the scientific founder of Navitor,[3] Raze Therapeutics,[4] and KSQ Therapeutics.[5]
Scientific contributions
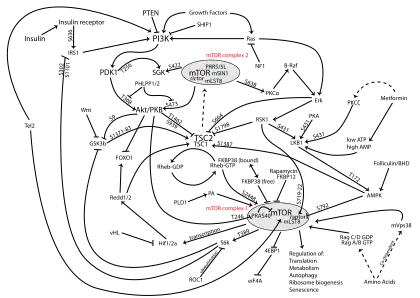
As a graduate student in Solomon Snyder’s Lab at Johns Hopkins, Sabatini began working on understanding the molecular mechanism of rapamycin; a macrolide antibiotic discovered in the soil of Easter Island that has potent antifungal, immunosuppressive, and anti-tumorigenic properties.[1] Although the TOR/DRR genes had been identified in 1993 as conferring rapymycin resistance in budding yeast, the direct target of rapamycin and its mechanism of action in mammals was unknown.[6][7] In 1994, Sabatini used rapamycin and its binding partner FKBP12 to purify the mechanistic Target of Rapamycin (mTOR) protein from rat brain, showing it to be the direct target of rapamycin in mammals and the homolog of the yeast TOR/DRR genes.[1]
Since starting his own lab at the Whitehead Institute in 1997, Sabatini has made numerous key contributions to the understanding of mTOR function, regulation, and importance in diseases such as cancer.[8] For example, his lab discovered the mTORC1[9] and mTORC2[10] multi-protein complexes, the nutrient sensing Rag GTPase pathway upstream of mTORC1,[11] as well as the direct amino acid sensors Sestrin[12][13] and CASTOR.[14][15]
Sabatini’s research interests have expanded in recent years to include cancer metabolism as well as technology development surrounding the use of high-throughput genetic screens in human cells, most notably through the use of RNA interference[16] and the CRISPR-Cas9 system.[17] As of 2016, Sabatini has authored over 250 publications and has an h-index of 100.
Selected awards and honors
- 2020 BBVA Foundation Frontiers of Knowledge Awards in Biology and Biomedicine
Selected publications
- Sabatini, DM (2017). "Twenty-five years of mTOR: Uncovering the link from nutrients to growth". Proc Natl Acad Sci U S A. 114 (45): 11818–11825. doi:10.1073/pnas.1716173114. PMC 5692607. PMID 29078414.
- Sabatini DM, Erdjument-Bromage H, Lui M, Tempst P, Snyder SH (1994). "RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs". Cell. 78 (1): 35–43. doi:10.1016/0092-8674(94)90570-3. PMID 7518356. S2CID 33647539.
References
- Viegas, Jennifer (26 December 2017). "Profile of David M. Sabatini". Proceedings of the National Academy of Sciences of the United States of America. 115 (3): 438–440. doi:10.1073/pnas.1721196115. PMC 5777013. PMID 29279397.
- Ware, Lauren (2013). "Science in their Blood". HHMI Bulletin. Howard Hughes Medical Institute. Retrieved 5 April 2017.
- "Navitor Pharmaceuticals". Archived from the original on 2017-11-14. Retrieved 2015-09-12.
- Raze Therapeutics
- Kendall Square Therapeutics
- Kunz J, Henriquez R, Schneider U, Deuter-Reinhard M, Movva NR, Hall MN (1993). "Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression". Cell. 73 (3): 585–96. doi:10.1016/0092-8674(93)90144-f. PMID 8387896. S2CID 42926249.
- Cafferkey R, Young PR, McLaughlin MM, Bergsma DJ, Koltin Y, Sathe GM, Faucette L, Eng WK, Johnson RK, Livi GP (1993). "Dominant missense mutations in a novel yeast protein related to mammalian phosphatidylinositol 3-kinase and VPS34 abrogate rapamycin cytotoxicity". Mol. Cell. Biol. 13 (10): 6012–23. doi:10.1128/mcb.13.10.6012. PMC 364661. PMID 8413204.
- Saxton RA, Sabatini, DM (2017). "mTOR Signaling in Growth, Metabolism, and Disease". Cell. 169 (2): 361–371. doi:10.1016/j.cell.2017.03.035. PMID 28388417.
- Kim DH, Sarbassov DD, Ali SM, et al. (2002). "mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery". Cell. 110 (2): 163–75. doi:10.1016/s0092-8674(02)00808-5. PMID 12150925. S2CID 4656930.
- Sarbassov DD, Ali SM, Kim DH, et al. (2004). "Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton". Curr. Biol. 14 (14): 1296–302. doi:10.1016/j.cub.2004.06.054. PMID 15268862. S2CID 4658268.
- Sancak Y, Peterson TR, Shaul YD, et al. (2008). "The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1". Science. 320 (5882): 1496–501. Bibcode:2008Sci...320.1496S. doi:10.1126/science.1157535. PMC 2475333. PMID 18497260.
- Wolfson RL, Chantranupong L, Saxton RA, et al. (2016). "Sestrin2 is a leucine sensor for the mTORC1 pathway". Science. 351 (6268): 43–8. Bibcode:2016Sci...351...43W. doi:10.1126/science.aab2674. PMC 4698017. PMID 26449471.
- Saxton RA, Knockenhauer KE, Wolfson RL, et al. (2016). "Structural basis for leucine sensing by the Sestrin2-mTORC1 pathway". Science. 351 (6268): 53–8. Bibcode:2016Sci...351...53S. doi:10.1126/science.aad2087. PMC 4698039. PMID 26586190.
- Chantranupong L, Scaria SM, Saxton RA, et al. (2016). "The CASTOR Proteins Are Arginine Sensors for the mTORC1 Pathway". Cell. 165 (1): 153–64. doi:10.1016/j.cell.2016.02.035. PMC 4808398. PMID 26972053.
- Saxton RA, Chantranupong L, Knockenhauer KE, Schwartz TU, Sabatini DM (2016). "Mechanism of arginine sensing by CASTOR1 upstream of mTORC1". Nature. 536 (7615): 229–33. Bibcode:2016Natur.536..229S. doi:10.1038/nature19079. PMC 4988899. PMID 27487210.
- Moffat J, Grueneberg DA, Yang X, et al. (2006). "A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen". Cell. 124 (6): 1283–98. doi:10.1016/j.cell.2006.01.040. PMID 16564017. S2CID 630641.
- Wang T, Wei JJ, Sabatini DM, Lander ES (2014). "Genetic screens in human cells using the CRISPR-Cas9 system" (PDF). Science. 343 (6166): 80–4. Bibcode:2014Sci...343...80W. doi:10.1126/science.1246981. PMC 3972032. PMID 24336569.
- Louisa Gross Horwitz Prize 2019