Dynamic speckle
In physics, dynamic speckle is a result of the temporal evolution of a speckle pattern where variations in the scattering elements responsible for the formation of the interference pattern in the static situation produce the changes that are seen in the speckle pattern, where its grains change their intensity (grey level) as well as their shape along time. One easy to observe example is milk: place some milk in a teaspoon and observe the surface in direct sunlight. There will be a visible "dancing" pattern of coloured points. Where the milk dries on the spoon at the edge, the speckle is seen to be static. This is direct evidence of the thermal motion of atoms, which cause the Brownian motion of the colloidal particles in the milk, which in turn results in the dynamic speckle visible to the naked eye.
Information content


The dynamic pattern shows then the changes that, if they are analyzed along time, represent the activity of the illuminated material. The visual effect is that of a boiling liquid or the image in a TV set far from tuning.
It can be analyzed by means of several mathematical and statistical tools and provide numeric or visual information on its magnitude, the not well defined idea of activity. Because the number of scattering centers is very high the collective phenomenon is hard to interpret and their individual contributions to the final result can not be inferred. The measurements that are obtained by means of the analysis tools present the activity level as a sum of the contributions of phenomena due to Doppler effect of the scattered light as well as other phenomena eventually present (time variations of the refractive index of the sample, etc.) Light scattered with small Doppler shifts in its frequency beats on the detector (eventually the eye) giving rise to the slow intensity variations that constitute the dynamic of the speckle pattern.
A biological sample, for example, that is a material that contains a huge number of mobile scattering centers, presents refractive index variations in the materials that compose it with power changes as well as many other effects increasing the complexity in the identification and isolation of these phenomena. Then, the complete interpretation of the activity of a sample, by means of dynamic speckle, presents itself big challenges.[1]
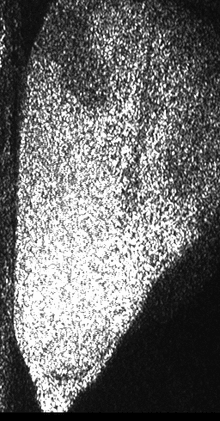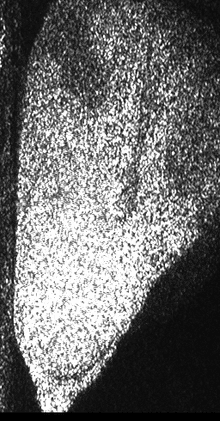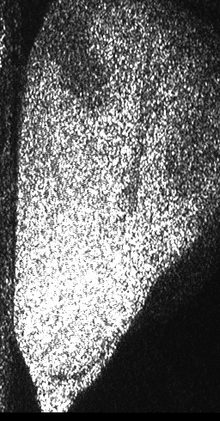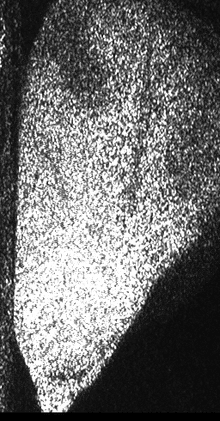
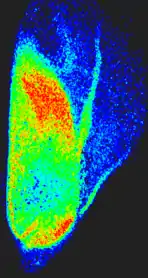
Figure 1 shows a sequence of speckle patterns in a corn seed in the start of its germination process where the dynamic effect is higher in the areas where the scattering centers are expected to be more active as is the case of the embryo and in a break in the endosperm region of the seed. The embryo is in the lower left side and the break is a river-like region in the center. In the crack, the activity is due to intensive inner water evaporation while in the embryo activity is higher due to metabolism of the alive tissue together with the activity caused by water evaporation. In the endosperm, the high right region of the image represents that the relatively low activity is due only to water evaporation.
Applications
Biological tissue is one of the most complex that can be found in nature. Besides it is worsened by the intrinsic variability present between one sample and another. These facts make even more difficult the comparison of results between different samples even in presence of the same stimulus. In this context, speckle patterns have been applied to study bacteria,[2][3] parasites, seeds and plants.[4]
Other fields of application are the analysis of drying paint,[5] control in gels,[6] foams, corrosion, efflorescence, etc.
Dynamic Speckle analysis
Several mathematical and statistical tools have been proposed for the characterization of the activity of a dynamic speckle pattern. Some of them are:
- Inertia Moment of the Co-Occurrence matrix (MOC)[7]
- Fujii[8]
- Generalized differences[9]
- Temporal difference[10]
These and other methods are gathered in Biospeckle laser tool library.
See also
References
- Rabal, HJ; Braga, RA (2008). Dynamic Laser Speckle and Applications. CRC Press. ISBN 978-1-4200-6015-7.
- Murialdo, S; et al. "Analysis of bacterial chemotactic response using dynamic laser speckle". J. Biomed. Opt. 14(6) (2009) 064015.
- Ramírez-Miquet, EE; et al. "Escherichia coli activity characterization using a laser dynamic speckle technique". Rev. Cub. Fis. 28(1E) (2011) pp. 1E13-1E17.
- Zhao, Y (1997). "Point-wise and whole-field laser speckle intensity fluctuation measurements applied to botanical specimens". Optics and Lasers in Engineering. 28 (6): 443–456. Bibcode:1997OptLE..28..443Z. doi:10.1016/S0143-8166(97)00056-0.
- Faccia, PA; et al. (2009). "Differentiation of the drying time of paints by dynamic speckle interferometry". Progress in Organic Coatings. 64 (4): 350–355. doi:10.1016/j.porgcoat.2008.07.016.
- Cabelo, CI; et al. Hydrophilic character study of silica-gel by a laser dynamic speckle method. Rev. Cub. Fis. 25(2A) (2008) pp. 67-69
- Arizaga, R (1999). "Speckle time evolution characterization by the co-occurrence matrix analysis". Optics & Laser Technology. 31 (2): 163–169. Bibcode:1999OptLT..31..163A. doi:10.1016/S0030-3992(99)00033-X.
- Briers, J (1995). "Quasi real-time digital version of single-exposure speckle photography for full-field monitoring of velocity or flow fields". Optics Communications. 116 (1–3): 36–42. Bibcode:1995OptCo.116...36B. doi:10.1016/0030-4018(95)00042-7.
- Arizaga, R.; et al. (2002). "Display of local activity using dynamical speckle patterns". Optical Engineering. 41 (2): 287. Bibcode:2002OptEn..41..287A. doi:10.1117/1.1428739.
- Martí-López, L.; et al. (2010). "Temporal difference method for processing dynamic speckle patterns". Optics Communications. 283 (24): 4972–4977. Bibcode:2010OptCo.283.4972M. doi:10.1016/j.optcom.2010.07.073.