ETFA
The human ETFA gene encodes the Electron-transfer-flavoprotein, alpha subunit, also known as ETF-α.[5] Together with Electron-transfer-flavoprotein, beta subunit, encoded by the 'ETFB' gene, it forms the heterodimericElectron transfer flavoprotein (ETF). The native ETF protein contains one molecule of FAD and one molecule of AMP, respectively.[6][7]
First reports on the ETF protein were based on ETF isolated from porcine liver.[8] Porcine and human ETF transfer electrons from mitochondrial matrix flavoenzymes to Electron transfer flavoprotein-ubiquinone oxidoreductase (ETF-QO) encoded by the ETFDH gene. ETF-QO subsequently relays the electrons via ubiquinone to complex III in the respiratory chain.[9] The flavoenzymes that transfer electrons to ETF are involved in fatty acid beta oxidation, amino acid catabolism, choline metabolism, and special metabolic pathways. Defects in either of the ETF subunits or ETFDH cause multiple acyl CoA dehydrogenase deficiency (OMIM # 231680),[10] earlier called glutaric acidemia type II. MADD is characterized by excretion of a series of substrates of the upstream flavoenzyes, e.g. glutaric, lactic, ethylmalonic, butyric, isobutyric, 2-methyl-butyric, and isovaleric acids.[5]
Evolutionary relationships
ETF is an evolutionarily ancient protein with orthologues found in all kingdoms of life. [11] ETFs are grouped into 3 subgroups, I, II, and III. The best studied group are group I ETFs that in eukaryotic cells are localized in the mitochondrial matrix space. Group I ETFs transfer electrons between flavoenzymes. Group II ETFs may also receive electrons from ferredoxin or NADH.[12]
Gene, expression, and subcellular localization
The human ETFA gene encoding the alpha subunit of ETF (ETF-α) is localized on chromosome 15 (15q24.2-q24.3). It is composed of 12 exons. Little is known about its promoter and transcriptional regulation. Global expression analyses show that it is expressed at substantial levels in most tissues (PROTEOMICXS DB). ETF-α is translated as a precursor protein with an N-terminal mitochondrial targeting sequence. [13] It is posttranslationally imported into the mitochondrial matrix space, where the targeting sequence is cut off.
Posttranslational modifications and regulation
Acetylation and succinylation of lysine residues and phosphorylation of serine and threonine residues in ETF-α have been reported in mass spectrometric analyses of posttranslational modifications P13804. Electron transfer flavoprotein regulatory factor 1 (ETFRF1) has been identified as a protein that specifically binds ETF and this interaction has been indicated to inactivate ETF by displacing the FAD.[14]
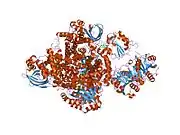
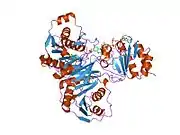
Structure and interaction with redox partners
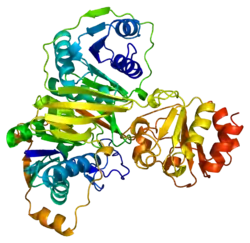
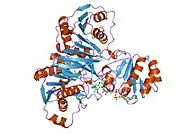
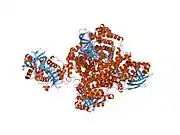
As first shown for porcine ETF, one chain of ETF-α assembles with one chain of ETF-β, and one molecule each of FAD and AMP to the dimeric native enzyme. [15][16][17][18] The crystal structure of human ETF was reported in 1996.[19] This showed that ETF consists of three distinct domains (I, II, and III). The FAD is bound in a cleft between the two subunits and interacts mainly with the C-terminal part of ETF-α. The AMP is buried in domain III. A crystal structure of the complex of one of its interactors, medium-chain acyl-CoA dehydrogenase (MCAD; gene name ACADM) has been determined.[20][21] (toogood 2004+2007). This identified a so-called recognition loop formed by ETF-β that anchors ETF on one subunit of the homotetrameric MCAD enzyme. This interaction triggers conformational changes and the highly mobile redox active FAD domain of ETF swings to the FAD domain of a neighboring subunit of the MCAD tetramer bringing the two FAD molecules into close contact for interprotein electron transfer.
Molecular Function
Human ETF receives electrons from at least 14 flavoenzymes and transfers them to ETF-ubiquinone oxidoreductases (ETF:QO) in the inner mitochondrial membrane. ETF:QO in turn relays them to ubiquinone from where they enter the respiratory chain at complex III. [22] Most of the flavoenzymes transferring electrons to ETF are participating in fatty acid oxidation, amino acid catabolism, and choline metabolism. ETF and ETF:QO thus represent an important hub for transfer of electrons from various redox reactions and feeding them into the respiratory chain for energy production.
Genetic deficiencies and molecular pathogenesis
Deleterious mutations in the ETFA and ETFB genes encoding ETF or the ETFDH gene encoding ETF:QO are associated with multiple acyl-CoA dehydrogenase deficiency (MADD; OMIM #231680; previously called glutaric aciduria type II).[23] Biochemically, MADD is characterized by elevated levels of a series of carnitine conjugates of the substrates of the different partner dehydrogenases of the ETF/ETF:QO hub, e.g. glutaric, lactic, ethylmalonic, butyric, isobutyric, 2-methyl-butyric, and isovaleric acids.[5] Accumulation of substrates and derivatives of the upstream dehydrogenases and energy deficiency upon fasting cause the clinical phenotype. Mostly depending on the severity of the mutation, the disease is divided into three subgroups: type I (neonatal onset with congenital anomalies), type II (neonatal onset without congenital anomalies), and type III (late onset). There is no cure for the disease, and treatment is employing a diet limiting protein and fat intake, avoidance of prolonged fasting, both to alleviate the flow through the partner dehydrogenases. In addition, supplementation of riboflavin, the precursor of the FAD co-factor can stabilize mutant ETF and ETF:QO variants with certain missense mutations.[24][25]
References
- GRCh38: Ensembl release 89: ENSG00000140374 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000032314 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Entrez Gene: ETFA electron-transfer-flavoprotein, alpha polypeptide (glutaric aciduria II)".
- Sato K, Nishina Y, Shiga K (August 1993). "Electron-transferring flavoprotein has an AMP-binding site in addition to the FAD-binding site". Journal of Biochemistry. 114 (2): 215–22. doi:10.1093/oxfordjournals.jbchem.a124157. PMID 8262902.
- Husain M, Steenkamp DJ (February 1983). "Electron transfer flavoprotein from pig liver mitochondria. A simple purification and re-evaluation of some of the molecular properties". The Biochemical Journal. 209 (2): 541–5. doi:10.1042/bj2090541. PMC 1154123. PMID 6847633.
- Crane FL, Beinert H (September 1954). "A Link Between Fatty Acyl CoA Dehydrogenase and Cytochrome C: A New Flavin Enzyme". Journal of the American Chemical Society. 76 (17): 4491. doi:10.1021/ja01646a076.
- Ruzicka FJ, Beinert H (December 1977). "A new iron-sulfur flavoprotein of the respiratory chain. A component of the fatty acid beta oxidation pathway". The Journal of Biological Chemistry. 252 (23): 8440–5. PMID 925004.
- "OMIM Entry - # 231680 - MULTIPLE ACYL-CoA DEHYDROGENASE DEFICIENCY; MADD". www.omim.org.
- Toogood HS, Leys D, Scrutton NS (November 2007). "Dynamics driving function: new insights from electron transferring flavoproteins and partner complexes". The FEBS Journal. 274 (21): 5481–504. doi:10.1111/j.1742-4658.2007.06107.x. PMID 17941859.
- Toogood HS, Leys D, Scrutton NS (November 2007). "Dynamics driving function: new insights from electron transferring flavoproteins and partner complexes". The FEBS Journal. 274 (21): 5481–504. doi:10.1111/j.1742-4658.2007.06107.x. PMID 17941859.
- Ikeda Y, Keese SM, Tanaka K (October 1986). "Biosynthesis of electron transfer flavoprotein in a cell-free system and in cultured human fibroblasts. Defect in the alpha subunit synthesis is a primary lesion in glutaric aciduria type II". The Journal of Clinical Investigation. 78 (4): 997–1002. doi:10.1172/JCI112691. PMC 423742. PMID 3760196.
- Floyd BJ, Wilkerson EM, Veling MT, Minogue CE, Xia C, Beebe ET, et al. (August 2016). "Mitochondrial Protein Interaction Mapping Identifies Regulators of Respiratory Chain Function". Molecular Cell. 63 (4): 621–632. doi:10.1016/j.molcel.2016.06.033. PMC 4992456. PMID 27499296.
- Hall CL, Kamin H (May 1975). "The purification and some properties of electron transfer flavoprotein and general fatty acyl coenzyme A dehydrogenase from pig liver mitochondria". The Journal of Biological Chemistry. 250 (9): 3476–86. PMID 1168197.
- Gorelick RJ, Mizzer JP, Thorpe C (December 1982). "Purification and properties of electron-transferring flavoprotein from pig kidney". Biochemistry. 21 (26): 6936–42. doi:10.1021/bi00269a049. PMID 7159575.
- Sato K, Nishina Y, Shiga K (August 1996). "In vitro refolding and unfolding of subunits of electron-transferring flavoprotein: characterization of the folding intermediates and the effects of FAD and AMP on the folding reaction". Journal of Biochemistry. 120 (2): 276–85. doi:10.1093/oxfordjournals.jbchem.a021410. PMID 8889811.
- Sato K, Nishina Y, Shiga K (August 1993). "Electron-transferring flavoprotein has an AMP-binding site in addition to the FAD-binding site". Journal of Biochemistry. 114 (2): 215–22. doi:10.1093/oxfordjournals.jbchem.a124157. PMID 8262902.
- Roberts DL, Frerman FE, Kim JJ (December 1996). "Three-dimensional structure of human electron transfer flavoprotein to 2.1-A resolution". Proceedings of the National Academy of Sciences of the United States of America. 93 (25): 14355–60. doi:10.1073/pnas.93.25.14355. PMC 26136. PMID 8962055.
- Toogood HS, van Thiel A, Basran J, Sutcliffe MJ, Scrutton NS, Leys D (July 2004). "Extensive domain motion and electron transfer in the human electron transferring flavoprotein.medium chain Acyl-CoA dehydrogenase complex". The Journal of Biological Chemistry. 279 (31): 32904–12. doi:10.1074/jbc.M404884200. PMID 15159392. S2CID 6901700.
- Toogood HS, Leys D, Scrutton NS (November 2007). "Dynamics driving function: new insights from electron transferring flavoproteins and partner complexes". The FEBS Journal. 274 (21): 5481–504. doi:10.1111/j.1742-4658.2007.06107.x. PMID 17941859.
- Ruzicka FJ, Beinert H (December 1977). "A new iron-sulfur flavoprotein of the respiratory chain. A component of the fatty acid beta oxidation pathway". The Journal of Biological Chemistry. 252 (23): 8440–5. PMID 925004.
- Prasun P (1993). Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJ, Stephens K, Amemiya A (eds.). "Multiple Acyl-CoA Dehydrogenase Deficiency". PMID 32550677. Cite journal requires
|journal=(help) - Henriques BJ, Olsen RK, Bross P, Gomes CM (2010). "Emerging roles for riboflavin in functional rescue of mitochondrial β-oxidation flavoenzymes". Current Medicinal Chemistry. 17 (32): 3842–54. doi:10.2174/092986710793205462. PMID 20858216.
- Henriques BJ, Bross P, Gomes CM (November 2010). "Mutational hotspots in electron transfer flavoprotein underlie defective folding and function in multiple acyl-CoA dehydrogenase deficiency" (PDF). Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease. 1802 (11): 1070–7. doi:10.1016/j.bbadis.2010.07.015. PMID 20674745.
Further reading
- Frerman FE (June 1988). "Acyl-CoA dehydrogenases, electron transfer flavoprotein and electron transfer flavoprotein dehydrogenase". Biochemical Society Transactions. 16 (3): 416–8. doi:10.1042/bst0160416. PMID 3053288.
- Freneaux E, Sheffield VC, Molin L, Shires A, Rhead WJ (November 1992). "Glutaric acidemia type II. Heterogeneity in beta-oxidation flux, polypeptide synthesis, and complementary DNA mutations in the alpha subunit of electron transfer flavoprotein in eight patients". The Journal of Clinical Investigation. 90 (5): 1679–86. doi:10.1172/JCI116040. PMC 443224. PMID 1430199.
- Indo Y, Glassberg R, Yokota I, Tanaka K (September 1991). "Molecular characterization of variant alpha-subunit of electron transfer flavoprotein in three patients with glutaric acidemia type II--and identification of glycine substitution for valine-157 in the sequence of the precursor, producing an unstable mature protein in a patient". American Journal of Human Genetics. 49 (3): 575–80. PMC 1683153. PMID 1882842.
- Finocchiaro G, Ito M, Ikeda Y, Tanaka K (October 1988). "Molecular cloning and nucleotide sequence of cDNAs encoding the alpha-subunit of human electron transfer flavoprotein". The Journal of Biological Chemistry. 263 (30): 15773–80. PMID 3170610.
- White RA, Dowler LL, Angeloni SV, Koeller DM (April 1996). "Assignment of Etfdh, Etfb, and Etfa to chromosomes 3, 7, and 13: the mouse homologs of genes responsible for glutaric acidemia type II in human". Genomics. 33 (1): 131–4. doi:10.1006/geno.1996.0170. PMID 8617498.
- Roberts DL, Frerman FE, Kim JJ (December 1996). "Three-dimensional structure of human electron transfer flavoprotein to 2.1-A resolution". Proceedings of the National Academy of Sciences of the United States of America. 93 (25): 14355–60. doi:10.1073/pnas.93.25.14355. PMC 26136. PMID 8962055.
- Bross P, Pedersen P, Winter V, Nyholm M, Johansen BN, Olsen RK, et al. (June 1999). "A polymorphic variant in the human electron transfer flavoprotein alpha-chain (alpha-T171) displays decreased thermal stability and is overrepresented in very-long-chain acyl-CoA dehydrogenase-deficient patients with mild childhood presentation". Molecular Genetics and Metabolism. 67 (2): 138–47. doi:10.1006/mgme.1999.2856. PMID 10356313.
- Jones M, Talfournier F, Bobrov A, Grossmann JG, Vekshin N, Sutcliffe MJ, Scrutton NS (March 2002). "Electron transfer and conformational change in complexes of trimethylamine dehydrogenase and electron transferring flavoprotein". The Journal of Biological Chemistry. 277 (10): 8457–65. doi:10.1074/jbc.M111105200. PMID 11756429.
- Olsen RK, Andresen BS, Christensen E, Bross P, Skovby F, Gregersen N (July 2003). "Clear relationship between ETF/ETFDH genotype and phenotype in patients with multiple acyl-CoA dehydrogenation deficiency". Human Mutation. 22 (1): 12–23. doi:10.1002/humu.10226. PMID 12815589.
- Kimura K, Wakamatsu A, Suzuki Y, Ota T, Nishikawa T, Yamashita R, et al. (January 2006). "Diversification of transcriptional modulation: large-scale identification and characterization of putative alternative promoters of human genes". Genome Research. 16 (1): 55–65. doi:10.1101/gr.4039406. PMC 1356129. PMID 16344560.
- Schiff M, Froissart R, Olsen RK, Acquaviva C, Vianey-Saban C (June 2006). "Electron transfer flavoprotein deficiency: functional and molecular aspects". Molecular Genetics and Metabolism. 88 (2): 153–8. doi:10.1016/j.ymgme.2006.01.009. PMID 16510302.
- Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M (November 2006). "Global, in vivo, and site-specific phosphorylation dynamics in signaling networks". Cell. 127 (3): 635–48. doi:10.1016/j.cell.2006.09.026. PMID 17081983. S2CID 7827573.
- Chiong MA, Sim KG, Carpenter K, Rhead W, Ho G, Olsen RK, Christodoulou J (2007). "Transient multiple acyl-CoA dehydrogenation deficiency in a newborn female caused by maternal riboflavin deficiency". Molecular Genetics and Metabolism. 92 (1–2): 109–14. doi:10.1016/j.ymgme.2007.06.017. PMID 17689999.