Equivalence group
An equivalence group is a set of unspecified cells that have the same developmental potential or ability to adopt various fates. Our current understanding suggests that equivalence groups are limited to cells of the same ancestry, also known as sibling cells.[1] Often, cells of an equivalence group adopt different fates from one another.[2]
Equivalence groups assume various potential fates in two general, non-mutually exclusive ways. One mechanism, induction, occurs when a signal originating from outside of the equivalence group specifies a subset of the naïve cells.[2] Another mode, known as lateral inhibition, arises when a signal within an equivalence group causes one cell to adopt a dominant fate while others in the group are inhibited from doing so.[3] In many examples of equivalence groups, both induction and lateral inhibition are used to define patterns of distinct cell types.
Cells of an equivalence group that do not receive a signal adopt a default fate. Alternatively, cells that receive a signal take on different fates.[2][4] At a certain point, the fates of cells within an equivalence group become irreversibly determined, thus they lose their multipotent potential. The following provides examples of equivalence groups studied in nematodes and ascidians.
Vulva Precursor Cell Equivalence Group
Introduction
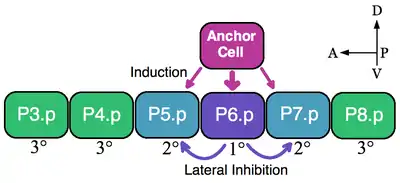
A classic example of an equivalence group is the vulva precursor cells (VPCs) of nematodes. In Caenorhabditis elegans self-fertilized eggs exit the body through the vulva. This organ develops from a subset of cell of an equivalence group consisting of six VPCs, P3.p-P8.p, which lie ventrally along the anterior-posterior axis.[5] In this example a single overlying somatic cells, the anchor cell, induces nearby VPCs to take on vulva fates 1° (P6.p) and 2° (P5.p and P7.p). VPCs that are not induced form the 3° lineage (P3.p, P4.p and P8.p), which make epidermal cells that fuse to a large syncytial epidermis (see image).[6]
The six VPCs form an equivalence group because all of the six cells are competent to take on any of the available fates (1°, 2°, and 3°) dependent on their proximity to the anchor cell. Ablation experiments indicate that all VPCs are able to adopt vulva fates. For example, if the P6.p cell that normally becomes 1° is ablated then the VPC closest to the anchor cell, either P5.p or P7.p, assumes the 1° fate. Furthermore, if all VPCs are destroyed except the most anterior P3.p cell then the anchor cell designates this cell the 1° fate. However, if the anchor cell is killed, in the absence of an inductive signal, then all of the VPCs assume the default 3° lineage.[7]
Molecular Mechanism
The anchor cell directly induces the vulva fates by secreting the epidermal growth factor (EGF)-like ligand LIN-3. The P6.p cell receives the LIN-3 signal via the receptor tyrosine kinase LET-23 (P5.p and P7.p also receive LIN-3 but to a lesser extent). Activation of LET-23 in P6.p results in the activation of LIN-12 (Notch) in P5.p and P7.p. Experimental evidence shows that LIN-12 is necessary and sufficient for the formation of the 2° fate. Through lateral inhibition LIN-12 prevents the P5.p and P7.p cells from adopting the 1° lineage.[7] Thus, in this example both inductive EGF signaling and lateral Notch activation patterns the VPC equivalence group.
Ascidian Pigment Precursor Equivalence Group
Introduction
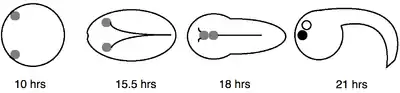
The larvae of ascidians (sea squirts) contain a pair of sensory pigment cells known as the otolith and ocellus. The otolith is used to sense gravity, whereas the ocellus responds to light. During embryogenesis the otolith and ocellus develop from two bilateral equivalent precursors. Either the left or right pigment precursor cell has equal probability of developing into the otolith or ocellus. The decision to adopt either fate is determined after neural tube closure during the early tailbud stage (see image), via a poorly defined mechanism of induction.[1]
During normal development, after neural tube closure, the pigment precursors align dorsally along the anterior-posterior axis of the neural tube. Whichever cell aligns anteriorly will become the otolith, while the posterior cell will form the ocellus. In the absence of cell-cell interactions both cells develop into ocelli, which is the default fate.[3][8]
Experimental Methods for Studying Equivalence in Halocynthia roretzi
To elucidate whether the fates of the otolith and ocellus are determined in the early embryo or after the precursors align during neural tube closure, ablation and drug treatment techniques were used in the ascidian species Halocythia roretzi.
Cells that are labeled with fluorescein isothiocyanate-dextran (FDX) can be selectively photoablated by fluorescent excitation.[9] When one FDX labeled pigment precursor cells is photoablated during the mid-neurula stage (15 hrs) the other will almost always develop into an ocellus. However, if the ablations are performed during the late tailbud stage (22.5 hrs) then the remaining cell has an equal likelihood of becoming an otolith or ocellus.[1]
Inhibiting cell division and morphogenesis with cytochalasin B is another method used to determine when the pigment precursor equivalence group is specified. Cytochalasin treatment of early tailbud stage embryos (17 hrs), while the two bilateral cells are still separated, results in both cells becoming ocelli. When the drug was used after the two cells aligned at the dorsal midline, the anterior cell developed into the otolith and the posterior cell became the ocellus without exception.[1] Both experiments suggest that fates of the pigment precursor cells are irreversibly determined by approximately the mid-tailbud stage (21 hrs).
Other Equivalence Groups
Equivalence groups have also been described in the ganglion mother cells in grasshopper and the O/P teloblasts in the leech.[10][11] Like other instances of equivalence groups, progeny cells are born equivalent and become specified through cell interactions. Equivalence groups are a common theme in the development of many organisms from diverse phyla.
References
- Nishida and Satoh; Satoh, N (1989). "Determination and regulation in the pigment cell lineage of the ascidian embryo". Dev Biol. 132 (2): 355–67. doi:10.1016/0012-1606(89)90232-7. PMID 2494088.
- Greenwald and Rubin; Rubin, GM (1992). "Making a difference: the role of cell-cell interactions in establishing separate identities for equivalent cells". Cell. 68 (2): 271–81. doi:10.1016/0092-8674(92)90470-W. PMID 1365402.
- Nishida; Saitoh, Takashi; Matsumoto, Midori; Makabe, Kazuhiro W.; Nishida, H. (1997). "Notch homologue from Halocynthia roretzi is preferentially expressed in the central nervous system during ascidian embryogenesis". Dev Genes Evol. 207 (6): 371–380. doi:10.1007/s004270050126. PMID 27747436.
- Huang and Weisblat; Weisblat, DA (1996). "Cell fate determination in an annelid equivalence group". Development. 122 (6): 1839–47. PMID 8674423.
- Kornfeld (1997). "Vulval development in Caenorhabditis elegans". Trends Genet. 13 (2): 55–61. doi:10.1016/S0168-9525(97)01005-6. PMID 9055606.
- Sternberg and Horvitz; Horvitz, HR (1986). "Pattern formation during vulval development in C. elegans". Cell. 44 (5): 761–72. doi:10.1016/0092-8674(86)90842-1. PMID 3753901.
- Sternberg, Paul W. (2005). "Vulval development". WormBook: 1–28. doi:10.1895/wormbook.1.6.1. PMC 4781130. PMID 18050418.
- Akanuma; et al. (2002). "Notch signaling is involved in nervous system formation in ascidian embryos". Development Genes and Evolution. 212 (10): 459–72. doi:10.1007/s00427-002-0264-x. PMID 12424517.
- Shankland and Weisblat; Weisblat, DA (1984). "Stepwise commitment of blast cell fates during the positional specification of the O and P cell lines in the leech embryo". Dev Biol. 106 (2): 326–42. doi:10.1016/0012-1606(84)90231-8. PMID 6500176.
- Kuwada and Goodman; Goodman, CS (1985). "Neuronal determination during embryonic development of the grasshopper nervous system". Dev Biol. 110 (1): 114–26. doi:10.1016/0012-1606(85)90069-7. PMID 4007260.
- Kuo and Shankland; Shankland, M (2004). "Evolutionary diversification of specification mechanisms within the O/P equivalence group of the leech genus Helobdella". Development. 131 (23): 5859–69. doi:10.1242/dev.01452. PMID 15525668.