Forensic DNA analysis
DNA profiling is the determination of a DNA profile for legal and investigative purposes. DNA analysis methods have changed numerous times over the years as technology improves and allows for more information to be determined with less starting material. Modern DNA analysis is based on the statistical calculation of the rarity of the produced profile within a population.
| Part of a series on |
| Forensic science |
|---|
 |
|
While most well known as a tool in forensic investigations, DNA profiling can also be used for non-forensic purposes. Paternity testing, and human genealogy research are just two of the non-forensic uses of DNA profiling.
History
The methods for producing a DNA profile were developed by Alec Jeffreys and his team in 1984.[1]
Methods
RFLP analysis

The first true method of DNA profiling was restriction fragment length polymorphism analysis. The first use of RFLP analysis in forensic casework was in 1985 in the United Kingdom.[2] This type of analysis used variable number tandem repeats (VNTRs) to distinguish between individuals. VNTRs are common throughout the genome and consist of the same DNA sequence repeated again and again.[3] Different individuals can have a different number of repeats at a specific location in the genome.[2] For example, person A could have four while person B could have 5 repeats. The differences were visualized through a process called gel electrophoresis. Smaller fragments would travel farther through the gel than larger fragments separating them out.[4] These differences were used to distinguish between individuals and when multiple VNTR sites were run together, RFLP analysis has a high degree of individualizing power.[5]
The process of RFLP analysis was extremely time consuming and due to the length of the repeats used, between 9 and 100 base pairs,[3][6] amplification methods such as the polymerase chain reaction could not be used. This limited RFLP to samples that already had a larger quantity of DNA available to start with and did not perform well with degraded samples.[7] RFLP analysis was the primary type of analysis performed in most forensic laboratories before finally being retired and replaced by newer methods. It was fully abandoned by the FBI in 2000 and replaced with STR analysis.[8]
DQ alpha testing
Developed in 1991,[8] DQ alpha testing was the first forensic DNA technique that utilized the polymerase chain reaction.[9] This technique allowed for the use of far fewer cells than RFLP analysis making it more useful for crime scenes that did not have the large amounts of DNA material that was previously required.[10] The DQ alpha 1 locus (or location) was also polymorphic and had multiple different alleles that could be used to limit the pool of individuals that could have produced that result and increasing the probability of exclusion.[11]
The DQ alpha locus was combined with other loci in a commercially available kit called Polymarker in 1993.[12] Polymarker was a precursor to modern multiplexing kits and allowed multiple different loci to be examined with one product. While more sensitive than RFLP analysis, Polymarker did not contain the same discriminatory power as the older RFLP testing.[12] By 1995, scientists attempted to return to a VNTR based analysis combined with PCR technology called amplified fragment length polymorphisms (AmpFLP).[8]
AmpFLP

AmpFLP was the first attempt to couple VNTR analysis with PCR for forensic casework. This method used shorter VNTRs than RFLP analysis, between 8 and 16 base pairs. The shorter base pair sizes of AmpFLP was designed to work better with the amplification process of PCR.[6] It was hoped that this technique would allow for the discriminating power of RFLP analysis with the ability to process samples that have less template DNA to work with or which were otherwise degraded. However, only a few loci were validated for forensic applications to work with AmpFLP analysis as forensic labs quickly moved on to other techniques limited its discriminating ability for forensic samples.[13]
The technique was ultimately never widely used although it is still in use in smaller countries due to its lower cost and simpler setup compared to newer methods.[14][15] By the late 1990s, laboratories began switching over to newer methods including STR analysis. These used even shorter fragments of DNA and could more reliably be amplified using PCR while still maintaining, and improving, the discriminatory power of the older methods.[8]
STR analysis
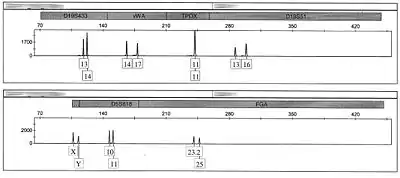
Short tandem repeat (STR) analysis is the primary type of forensic DNA analysis performed in modern DNA laboratories. STR analysis builds upon RFLP and AmpFLP used in the past by shrinking the size of the repeat units, to 2 to 6 base pairs, and by combining multiple different loci into one PCR reaction. These multiplexing assay kits can produce allele values for dozens of different loci throughout the genome simultaneously limiting the amount of time it takes to gain a full, individualizing, profile. STR analysis has become the gold standard for DNA profiling and is used extensively in forensic applications.
STR analysis can also be restricted to just the Y chromosome. Y-STR analysis can be used in cases that involve paternity or in familial searching as the Y chromosome is identical down the paternal line (except in cases where a mutation occurred). Certain multiplexing kits combine both autosomal and Y-STR loci into one kit further reducing the amount of time it takes to obtain a large amount of data.
mtDNA sequencing
Mitochondrial DNA sequencing is a specialized technique that uses the separate mitochondrial DNA present in most cells. This DNA is passed down the material line and is not unique between individuals. However, because of the number of mitochondria present in cells, mtDNA analysis can be used for highly degraded samples or samples where STR analysis would not produce enough data to be useful. mtDNA is also present in locations where autosomal DNA would be absent, such as in the shafts of hair.
Because of the increased chance of contamination when dealing with mtDNA, few laboratories process mitochondrial samples. Those that do have specialized protocols in place that further separate different samples from each other to avoid cross-contamination.
Rapid DNA
Rapid DNA is a "swab in-profile out" technology that completely automates the entire DNA extraction, amplification, and analysis process. Rapid DNA instruments are able to go from a swab to a DNA profile in as little as 90 minutes and eliminates the need for trained scientists to perform the process. These instruments are being looked at for use in the offender booking process allowing police officers to obtain the DNA profile of the person under arrest.
Recently, the Rapid DNA Act of 2017 was passed in the United States, directing the FBI to create protocols for the implementation of this technology throughout the country. Currently, DNA obtained from these instruments is not eligible for upload to national DNA databases as they do not analyze enough loci to meet the standard threshold. However, multiple police agencies already use Rapid DNA instruments to collect samples from people arrested in their area. These local DNA database are not subject to federal or state regulations.
Massively parallel sequencing
Also known as next-generation sequencing, massively parallel sequencing (MPS) builds upon STR analysis by introducing direct sequencing of the loci. Instead of the number of repeats present at each location, MPS would give the scientist the actual base pair sequence. Theoretically MPS has the ability to distinguish between identical twins as random point mutations would be seen within repeat segments that would not be picked up by traditional STR analysis.
Profile rarity
When a DNA profile is used in an evidentiary manner a match statistic is provided that explains how rare a profile is within a population. Specifically, this statistic is the probability that a person picked randomly out of a population would have that specific DNA profile. It is not the probability that the profile "matches" someone. There are multiple different methods to determining this statistic and each are used by various laboratories based on their experience and preference. However, likelihood ratio calculations is becoming the preferred method over the other two most commonly used methods, random man not excluded and combined probability of inclusion. Match statistics are especially important in mixture interpretation where there is more than one contributor to a DNA profile. When these statistics are given in a courtroom setting or in a laboratory report they are usually given for the three most common races of that specific area. This is because the allele frequencies at different loci changed based on the individual's ancestry. https://strbase.nist.gov/training/6_Mixture-Statistics.pdf
Random man not excluded
The probability produced with this method is the probability that a person randomly selected out the population could not be excluded from the analyzed data. This type of match statistic is easy to explain in a courtroom setting to individuals who have no scientific background but it also loses a lot of discriminating power as it does not take into account the suspect's genotype. This approach is commonly used when the sample is degraded or contains so many contributors that a singular profile cannot be determined. It is also useful in explaining to laypersons as the method of obtaining the statistic is straightforward. However, due to its limited discriminating power, RMNE is not generally performed unless no other method can be used. RMNE is not recommended for use in data that indicates a mixture is present.
Combined probability of inclusion/exclusion
.jpg.webp)
.jpg.webp)
Combined probability of inclusion or exclusion calculates the probability that a random, unrelated, person would be a contributor to a DNA profile or DNA mixture. In this method, statistics for each individual locus is determined using population statistics and then combined to get the total CPI or CPE. These calculations are repeated for all available loci with all available data and then each value is multiplied together to get the total combined probability of inclusion or exclusion. Since the values are multiplied together, extremely small numbers can be achieved using CPI. CPI or CPE is considered an acceptable statistical calculation when a mixture is indicated. https://www.promega.com/-/media/files/resources/conference-proceedings/ishi-15/parentage-and-mixture-statistics-workshop/generalpopulationstats.pdf?la=en
Example calculation for single source profile
Probability of a Caucasian having a 16 allele at vWA = .10204
Probability of a Caucasian having a 17 allele at vWA = .26276
Probability of a Caucasian having both a 14 and a 17 allele (P) = .10204 + .26276 = .3648
Probability of all other alleles being present (Q) = 1 - P or 1 - .3648 = .6352
Probability of exclusion for vWA = Q2 + 2Q(1-Q) or .63522 + 2(.6352)(1 - .6352) = .86692096 ≈ 86.69%
Probability of inclusion for vWA = 1 - CPE or 1 - .86692096 = .13307904 ≈ 13.31%
Example calculation for mixture profile
Probability of a Caucasian having a 14 allele at vWA = .10204
Probability of a Caucasian having a 15 allele at vWA = .11224
Probability of a Caucasian having a 16 allele at vWA = .20153
Probability of a Caucasian having a 19 allele at vWA = .08418
Probability of a Caucasian having a 14, 15, 16, or 19 allele (P) = .10204 + .11224 + .20153 + .08418 = .49999
Probability of all other alleles being present (Q) = 1 - P or 1 - .49999 = .50001
Probability of exclusion for vWA = Q2 + 2Q(1-Q) or .500012 + 2(.50001)(1 - .50001) = .7500099999 ≈ 75%
Probability of inclusion for vWA = 1 - CPE or 1 - .7500099999 = .2499900001 ≈ 25%
Likelihood ratio
Likelihood ratios (LR) are a comparison of two different probabilities to determine which one is more likely. When it involves a trial the LR is the probability of the prosecution's argument versus the probability of the defense's argument given their starting assumptions. In this scenario the prosecution's probability is often equal to 1 since the assumption is that the prosecution would not prosecute a suspect unless they were absolutely certain (100%) that they have the right person. Likelihood ratios are becoming more common in laboratories due to their usefulness in presenting statistics for data that indicates multiple contributors as well as their use in probabilistic genotyping software that predicts the most likely allele combinations given a set of data.
The drawbacks with using likelihood ratios is that they are very difficult to understand how analysts arrived at a specific value and the mathematics involved get very complicated as more data is introduced to the equations. In order to combat these problems in a courtroom setting, some laboratories have set up a "verbal scale" that replaces the actual numeral value of the likelihood ratio.
References
- McKie, Robin (May 23, 2009). "Eureka moment that led to the discovery of DNA fingerprinting". The Guardian. Retrieved October 29, 2017.
- "DNA Typing by RFLP Analysis". National Forensic Science Technology Center. 2005. Archived from the original on January 3, 2015. Retrieved November 10, 2017.
- Griffiths, Anthony J.F.; Lewontin, Richard C.; Gelbart, William M.; Miller, Jeffrey H. Modern Genetic Analysis: Integrating Genes and Genomes (Second ed.). W. H. Freeman and Company. p. 274. Retrieved November 10, 2017.
- Fisher, Barry A. J. (2005). Techniques of Crime Scene Investigation (Seventh ed.). CRC Press. p. 240. Retrieved November 11, 2017.
- Rudin, Norah; Inman, Keith (2002). An Introduction to Forensic DNA Analysis (Second ed.). CRC Press. p. 41. Retrieved November 11, 2017.
- James, Stuart H.; Nordby, Jon J., eds. (2005). Forensic Science: An Introduction to Scientific and Investigative Techniques (Second ed.). Taylor & Francis. p. 286. Retrieved November 10, 2017.
- "Disadvantages". National Forensic Science Technology Center. 2005. Archived from the original on January 2, 2015. Retrieved November 10, 2017.
- Tilstone, William J.; Savage, Kathleen A.; Clark, Leigh A. (2006). Forensic Science: An Encyclopedia of History, Methods, and Techniques. ABC CLIO. p. 49. Retrieved October 30, 2017.
- Riley, Donald E. (April 6, 2005). "DNA Testing: An Introduction For Non-Scientists An Illustrated Explanation". Retrieved October 29, 2017.
- McClintock, J. Thomas (2008). Forensic DNA Analysis: A Laboratory Manual. CRC Press. p. 64. Retrieved October 29, 2017.
- Blake, E; Mihalovich, J; Hiquchi, R; Walsh, PS; Erlich, H (May 1992). "Polymerase chain reaction (PCR) amplification and human leukocyte antigen (HLA)-DQ alpha oligonucleotide typing on biological evidence samples: casework experience". Journal of Forensic Sciences. 37 (3): 700–726. PMID 1629670.
- "DQ-Alpha". National Forensic Science Technology Center. 2005. Archived from the original on November 10, 2014. Retrieved October 30, 2017.
- Bär, W.; Fiori, A.; Rossi, U., eds. (October 1993). Advances in Forensic Haemogenetics. 15th Congress of the International Society for Forensic Haemogenetics. p. 255. Retrieved November 10, 2017.
- "AmpFLPs". National Forensic Science Technology Center. 2005. Archived from the original on November 21, 2014. Retrieved November 5, 2017.
- "DNA Fingerprinting Methods". Fingerprinting.com. Retrieved November 5, 2017.