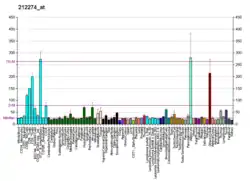
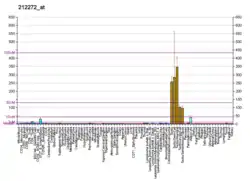
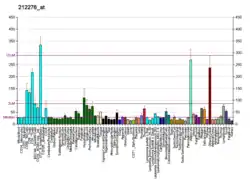
LPIN1
Lipin-1 is a protein that in humans is encoded by the LPIN1 gene.[5][6][7][8]
Function
Lipin-1 has phosphatidate phosphatase activity.[9][10][11] The nuclear localization of Lipin 1 is regulated by the mammalian Target Of Rapamycin protein kinase and links mTORC1 activity to the regulation of Sterol regulatory element-binding proteins (SREBP)-dependent gene transcription.[12][13][14]
Clinical significance
This gene represents a candidate gene for human lipodystrophy, characterized by loss of body fat, fatty liver, hypertriglyceridemia, and insulin resistance. Mouse studies suggest that this gene functions during normal adipose tissue development and may also play a role in human triglyceride metabolism.[8][12]
References
- GRCh38: Ensembl release 89: ENSG00000134324 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000020593 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Péterfy M, Phan J, Xu P, Reue K (January 2001). "Lipodystrophy in the fld mouse results from mutation of a new gene encoding a nuclear protein, lipin". Nature Genetics. 27 (1): 121–4. doi:10.1038/83685. PMID 11138012. S2CID 35168517.
- Nagase T, Seki N, Ishikawa K, Tanaka A, Nomura N (February 1996). "Prediction of the coding sequences of unidentified human genes. V. The coding sequences of 40 new genes (KIAA0161-KIAA0200) deduced by analysis of cDNA clones from human cell line KG-1". DNA Research. 3 (1): 17–24. doi:10.1093/dnares/3.1.17. PMID 8724849.
- Finck BN, Gropler MC, Chen Z, Leone TC, Croce MA, Harris TE, Lawrence JC, Kelly DP (September 2006). "Lipin 1 is an inducible amplifier of the hepatic PGC-1alpha/PPARalpha regulatory pathway". Cell Metabolism. 4 (3): 199–210. doi:10.1016/j.cmet.2006.08.005. PMID 16950137.
- "Entrez Gene: LPIN1 lipin 1".
- Han GS, Wu WI, Carman GM (April 2006). "The Saccharomyces cerevisiae Lipin homolog is a Mg2+-dependent phosphatidate phosphatase enzyme". The Journal of Biological Chemistry. 281 (14): 9210–8. doi:10.1074/jbc.M600425200. PMC 1424669. PMID 16467296.
- Donkor J, Sariahmetoglu M, Dewald J, Brindley DN, Reue K (February 2007). "Three mammalian lipins act as phosphatidate phosphatases with distinct tissue expression patterns". The Journal of Biological Chemistry. 282 (6): 3450–7. doi:10.1074/jbc.M610745200. PMID 17158099.
- Harris TE, Huffman TA, Chi A, Shabanowitz J, Hunt DF, Kumar A, Lawrence JC (January 2007). "Insulin controls subcellular localization and multisite phosphorylation of the phosphatidic acid phosphatase, lipin 1". The Journal of Biological Chemistry. 282 (1): 277–86. doi:10.1074/jbc.M609537200. PMID 17105729.
- Peterson TR, Sengupta SS, Harris TE, Carmack AE, Kang SA, Balderas E, Guertin DA, Madden KL, Carpenter AE, Finck BN, Sabatini DM (August 2011). "mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway". Cell. 146 (3): 408–20. doi:10.1016/j.cell.2011.06.034. PMC 3336367. PMID 21816276.
- Kennedy BK, Lamming DW (June 2016). "The Mechanistic Target of Rapamycin: The Grand ConducTOR of Metabolism and Aging". Cell Metabolism. 23 (6): 990–1003. doi:10.1016/j.cmet.2016.05.009. PMC 4910876. PMID 27304501.
- Laplante M, Sabatini DM (April 2013). "Regulation of mTORC1 and its impact on gene expression at a glance". Journal of Cell Science. 126 (Pt 8): 1713–9. doi:10.1242/jcs.125773. PMC 3678406. PMID 23641065.
Further reading
- Péterfy M, Phan J, Oswell GM, Xu P, Reue K (December 1999). "Genetic, physical, and transcript map of the fld region on mouse chromosome 12". Genomics. 62 (3): 436–44. doi:10.1006/geno.1999.6023. PMID 10644441.
- Reue K, Xu P, Wang XP, Slavin BG (July 2000). "Adipose tissue deficiency, glucose intolerance, and increased atherosclerosis result from mutation in the mouse fatty liver dystrophy (fld) gene". Journal of Lipid Research. 41 (7): 1067–76. PMID 10884287.
- Huffman TA, Mothe-Satney I, Lawrence JC (January 2002). "Insulin-stimulated phosphorylation of lipin mediated by the mammalian target of rapamycin". Proceedings of the National Academy of Sciences of the United States of America. 99 (2): 1047–52. doi:10.1073/pnas.022634399. PMC 117427. PMID 11792863.
- Cao H, Hegele RA (2003). "Identification of single-nucleotide polymorphisms in the human LPIN1 gene". Journal of Human Genetics. 47 (7): 370–2. doi:10.1007/s100380200052. PMID 12111372.
- Suviolahti E, Reue K, Cantor RM, Phan J, Gentile M, Naukkarinen J, Soro-Paavonen A, Oksanen L, Kaprio J, Rissanen A, Salomaa V, Kontula K, Taskinen MR, Pajukanta P, Peltonen L (February 2006). "Cross-species analyses implicate Lipin 1 involvement in human glucose metabolism". Human Molecular Genetics. 15 (3): 377–86. doi:10.1093/hmg/ddi448. PMID 16357106.
- Yao-Borengasser A, Rasouli N, Varma V, Miles LM, Phanavanh B, Starks TN, Phan J, Spencer HJ, McGehee RE, Reue K, Kern PA (October 2006). "Lipin expression is attenuated in adipose tissue of insulin-resistant human subjects and increases with peroxisome proliferator-activated receptor gamma activation". Diabetes. 55 (10): 2811–8. doi:10.2337/db05-1688. PMID 17003347.
- van Harmelen V, Rydén M, Sjölin E, Hoffstedt J (January 2007). "A role of lipin in human obesity and insulin resistance: relation to adipocyte glucose transport and GLUT4 expression". Journal of Lipid Research. 48 (1): 201–6. doi:10.1194/jlr.M600272-JLR200. PMID 17035674.
- Harris TE, Huffman TA, Chi A, Shabanowitz J, Hunt DF, Kumar A, Lawrence JC (January 2007). "Insulin controls subcellular localization and multisite phosphorylation of the phosphatidic acid phosphatase, lipin 1". The Journal of Biological Chemistry. 282 (1): 277–86. doi:10.1074/jbc.M609537200. PMID 17105729.
- Croce MA, Eagon JC, LaRiviere LL, Korenblat KM, Klein S, Finck BN (September 2007). "Hepatic lipin 1beta expression is diminished in insulin-resistant obese subjects and is reactivated by marked weight loss". Diabetes. 56 (9): 2395–9. doi:10.2337/db07-0480. PMID 17563064.
This article is issued from Wikipedia. The text is licensed under Creative Commons - Attribution - Sharealike. Additional terms may apply for the media files.