Lysine riboswitch
The Lysine riboswitch is a metabolite binding RNA element found within certain messenger RNAs that serve as a precision sensor for the amino acid lysine. Allosteric rearrangement of mRNA structure is mediated by ligand binding, and this results in modulation of gene expression.[1] Lysine riboswitch are most abundant in Firmicutes and Gammaproteobacteria where they are found upstream of a number of genes involved in lysine biosynthesis, transport and catabolism.[2][3][4] The lysine riboswitch has also been identified independently and called the L box.[5]
| Lysine riboswitch | |
|---|---|
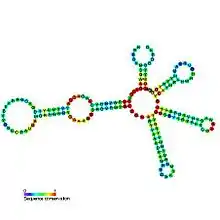 Predicted secondary structure and sequence conservation of Lysine riboswitch | |
| Identifiers | |
| Symbol | Lysine |
| Rfam | RF00168 |
| Other data | |
| RNA type | Cis-reg; riboswitch |
| Domain(s) | Bacteria |
| SO | SO:0000035 |
| PDB structures | PDBe |
Structure
The structure of the lysine riboswitch has recently been determined.[6][7] The lysine amino acid is bound in the pocket formed by the 5-way junction. The structure is composed of a three helical bundle and a two helical bundle joined by the 5-way junction. Helices 1 and 2 are stacked in a colinear fashion as are helices 4 and 5.
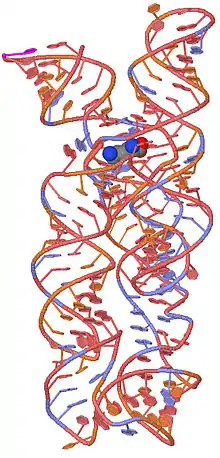 A 3D representation of the lysine riboswitch with a bound lysine molecule shown in space filling spheres.[6]
A 3D representation of the lysine riboswitch with a bound lysine molecule shown in space filling spheres.[6]
References
- Mandal, M; Boese B; Barrick JE; Winkler WC; Breaker RR (2003). "Riboswitches Control Fundamental Biochemical Pathways in Bacillus subtilis and Other Bacteria". Cell. 113 (5): 577–586. doi:10.1016/S0092-8674(03)00391-X. PMID 12787499. S2CID 8012149.
- Sudarsan, N; Wickiser JK; Nakamura S; Ebert MS; Breaker RR (2003). "An mRNA structure in bacteria that controls gene expression by binding lysine". Genes Dev. 17 (21): 2688–2697. doi:10.1101/gad.1140003. PMC 280618. PMID 14597663.
- Rodionov, DA; Vitreschak AG; Mironov AA; Gelfand MS (2003). "Regulation of lysine biosynthesis and transport genes in bacteria: yet another RNA riboswitch?". Nucleic Acids Res. 31 (23): 6748–6757. doi:10.1093/nar/gkg900. PMC 290268. PMID 14627808.
- Mukherjee, S; Barash D; Sengupta S (2017). "Comparative genomics and phylogenomic analyses of lysine riboswitch distributions in bacteria". PLOS ONE. 12 (9): e0184314. doi:10.1371/journal.pone.0184314. PMC 5584792. PMID 28873470.
- Grundy, FJ; Lehman SC; Henkin TM (2003). "The L box regulon: Lysine sensing by leader RNAs of bacterial lysine biosynthesis genes". Proc Natl Acad Sci USA. 100 (21): 12057–12062. doi:10.1073/pnas.2133705100. PMC 218712. PMID 14523230.
- Serganov A, Huang L, Patel DJ (2008). "Structural insights into amino acid binding and gene control by a lysine riboswitch". Nature. 455 (7217): 1263–1267. doi:10.1038/nature07326. PMC 3726722. PMID 18784651.
- Garst AD, Héroux A, Rambo RP, Batey RT (August 2008). "Crystal structure of the lysine riboswitch regulatory mRNA element". The Journal of Biological Chemistry. 283 (33): 22347–22351. doi:10.1074/jbc.C800120200. PMC 2504901. PMID 18593706.