Noninvasive genotyping
Noninvasive genotyping is a modern technique for obtaining DNA for genotyping that is characterized by the indirect sampling of specimen, not requiring harm to, handling of, or even the presence of the organism of interest. Beginning in the early 1990s, with the advent of PCR, researchers have been able to obtain high-quality DNA samples from small quantities of hair, feathers, scales, or excrement. These noninvasive samples are an improvement over older allozyme and DNA sampling techniques that often required larger samples of tissue or the destruction of the studied organism. Noninvasive genotyping is widely utilized in conservation efforts, where capture and sampling may be difficult or disruptive to behavior.[1] Additionally, in medicine, this technique is being applied in humans for the diagnosis of genetic disease and early detection of tumors. In this context, invasivity takes on a separate definition where noninvasive sampling also includes simple blood samples.
Uses and Methods
Conservation
In conservation, noninvasive genotyping has been used to supplement traditional techniques with broadly ranging levels of success. Modern DNA amplification methods allow researchers to use fecal or hair samples collected from the field to assess basic information about the specimen, including sex or species.[2] Despite the potential that noninvasive genotyping has in conservation genetics efforts, the efficiency of this method is in question, as field samples often suffer from degradation and contamination or are difficult to procure.[3] For instance, a team of researchers successfully used coyote fecal samples to estimate the abundance of a population in Georgia, thereby avoiding the substantial difficulty and consequences involved in trapping and procuring samples from the animals.[2]
Fetal Genotyping
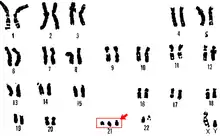
The most common use of noninvasive genotyping in medicine is non-invasive prenatal diagnosis (NIPD), which provides an alternative to riskier techniques such as amniocentesis. With the discovery of cell-free fetal DNA in maternal plasma, NIPD became a popular method for determining sex, paternity, aneuploidy, and the occurrence of monogenic diseases as it requires only a simple blood sample.[4][5] One NIPD provider maintains that a 10 mL blood sample will provide 99% accurate detection of basic genomic abnormalities as early as 10 weeks into pregnancy.[6] The karyotype below is that of an individual with trisomy 21, or Down Syndrome, which is what is most routinely checked for by NIPD screens.
Tumor Detection
This same technique is also utilized to identify the incidence of tumor DNA in the blood, which can both provide early detection of tumor growth and indicate relapse in cancer. Circulating tumor DNA can be found in the blood before metastasis occurs and, therefore, detection of certain mutant alleles may enhance survival rates in cancer patients.[7][8] In a recent study, ctDNA was shown to be "a broadly applicable, sensitive, and specific biomarker that can be used for a variety of clinical and research purposes in patients with multiple different types of cancer".[9] This technique is often referred to as a liquid biopsy, and has not been widely implemented in clinical settings although its impact could be quite large.[10] Although blood-borne ctDNA remains the most clinically significant noninvasive cancer detection, other studies have emerged that investigate other potential methods, including detection of colorectal cancer via fecal samples.[11]
Methods
The method by which samples are collected in noninvasive genotyping is what separates the technique from traditional genotyping, and there are a number of ways that this is accomplished. In the field, procured samples of tissue are captured, the tissue is dissolved, and the DNA is purified, although the exact procedure differs between different samples.[12] Following the collection of DNA samples, PCR technology is utilized to amplify particular genetic sequences, with PCR primer specificity avoiding contamination from other DNA sources. Then, the DNA can be analyzed using a number of genomic techniques, similarly to traditionally obtained samples.
References
- Woodruff, David S. "Noninvasive genotyping and field studies of free-ranging nonhuman primates." Kinship and behavior in primates (2004): 46-68.
- Gulsby, William D.; Killmaster, Charlie H.; Bowers, John W.; Laufenberg, Jared S.; Sacks, Benjamin N.; Statham, Mark J.; Miller, Karl V. (2016-12-01). "Efficacy and precision of fecal genotyping to estimate coyote abundance". Wildlife Society Bulletin. 40 (4): 792–799. doi:10.1002/wsb.712. ISSN 1938-5463.
- Anderson, Heather; McCafferty, Dominic; Saccheri, Ilik; McCluskie, Alan (2006-09-07). "Non-invasive genetic sampling of the Eurasian Otter ( Lutra lutra ) using hairs". Hystrix, the Italian Journal of Mammalogy. 17 (1). doi:10.4404/hystrix-17.1-4365. ISSN 1825-5272.
- Fan, H. Christina; Gu, Wei; Wang, Jianbin; Blumenfeld, Yair J.; El-Sayed, Yasser Y.; Quake, Stephen R. (2012). "Non-invasive prenatal measurement of the fetal genome". Nature. 487 (7407): 320–324. doi:10.1038/nature11251. PMC 3561905. PMID 22763444.
- Lun, Fiona M. F.; Tsui, Nancy B. Y.; Chan, K. C. Allen; Leung, Tak Y.; Lau, Tze K.; Charoenkwan, Pimlak; Chow, Katherine C. K.; Lo, Wyatt Y. W.; Wanapirak, Chanane (2008-01-01). "Noninvasive Prenatal Diagnosis of Monogenic Diseases by Digital Size Selection and Relative Mutation Dosage on DNA in Maternal Plasma". Proceedings of the National Academy of Sciences of the United States of America. 105 (50): 19920–19925. doi:10.1073/pnas.0810373105. JSTOR 25465749. PMC 2596743. PMID 19060211.
- "NIFTY test methodology and sequencing technology". The NIFTY™ Test - A Non-Invasive Prenatal Test Brought To By BGI Diagnostics. Retrieved 2017-04-10.
- Scherer, Florian; Kurtz, David M.; Newman, Aaron M.; Stehr, Henning; Liu, Chih Long; Zhou, Li; Craig, Alexander F. M.; Chabon, Jacob J.; Lovejoy, Alexander F. (2015-12-03). "Noninvasive Genotyping and Assessment of Treatment Response in Diffuse Large B Cell Lymphoma". Blood. 126 (23): 114. ISSN 0006-4971.
- Siravegna, Giulia; Bardelli, Alberto (2016-03-01). "Blood circulating tumor DNA for non-invasive genotyping of colon cancer patients". Molecular Oncology. Thematic Issue: Liquid biopsies. 10 (3): 475–480. doi:10.1016/j.molonc.2015.12.005. PMC 5528968. PMID 26774880.
- Bettegowda, Chetan; Sausen, Mark; Leary, Rebecca J.; Kinde, Isaac; Wang, Yuxuan; Agrawal, Nishant; Bartlett, Bjarne R.; Wang, Hao; Luber, Brandon (2014-02-19). "Detection of Circulating Tumor DNA in Early- and Late-Stage Human Malignancies". Science Translational Medicine. 6 (224): 224ra24. doi:10.1126/scitranslmed.3007094. ISSN 1946-6234. PMC 4017867. PMID 24553385.
- Standaert, Michael. "Blood Test for Early Cancer Detection". MIT Technology Review. Retrieved 2017-04-10.
- Spethmann, Sebastian; Fischer, Carsten; Wagener, Christoph; Streichert, Thomas; Tschentscher, Peter (2004-03-01). "Nucleic acids from intact epithelial cells as a target for stool-based molecular diagnosis of colorectal cancer". International Journal of Molecular Medicine. 13 (3): 451–454. doi:10.3892/ijmm.13.3.451. ISSN 1107-3756. PMID 14767578.
- Ghatak, Souvik; Muthukumaran, Rajendra Bose; Nachimuthu, Senthil Kumar (2017-04-10). "A Simple Method of Genomic DNA Extraction from Human Samples for PCR-RFLP Analysis". Journal of Biomolecular Techniques. 24 (4): 224–231. doi:10.7171/jbt.13-2404-001. ISSN 1524-0215. PMC 3792701. PMID 24294115.