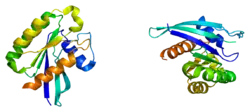RAB21
Ras-related protein Rab-21 is a protein that in humans is encoded by the RAB21 gene.[5][6][7][8]
References
- GRCh38: Ensembl release 89: ENSG00000080371 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000020132 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Opdam FJ, Kamps G, Croes H, van Bokhoven H, Ginsel LA, Fransen JA (Oct 2000). "Expression of Rab small GTPases in epithelial Caco-2 cells: Rab21 is an apically located GTP-binding protein in polarised intestinal epithelial cells". Eur J Cell Biol. 79 (5): 308–16. doi:10.1078/S0171-9335(04)70034-5. PMID 10887961.
- Pereira-Leal JB, Seabra MC (Nov 2001). "Evolution of the Rab family of small GTP-binding proteins". J Mol Biol. 313 (4): 889–901. doi:10.1006/jmbi.2001.5072. PMID 11697911.
- Pellinen T, Arjonen A, Vuoriluoto K, Kallio K, Fransen JA, Ivaska J (Jun 2006). "Small GTPase Rab21 regulates cell adhesion and controls endosomal traffic of beta1-integrins". J Cell Biol. 173 (5): 767–80. doi:10.1083/jcb.200509019. PMC 2063892. PMID 16754960.
- "Entrez Gene: RAB21 RAB21, member RAS oncogene family".
Further reading
- Nagase T, Miyajima N, Tanaka A, et al. (1995). "Prediction of the coding sequences of unidentified human genes. III. The coding sequences of 40 new genes (KIAA0081-KIAA0120) deduced by analysis of cDNA clones from human cell line KG-1". DNA Res. 2 (1): 37–43. doi:10.1093/dnares/2.1.37. PMID 7788527.
- Bao S, Zhu J, Garvey WT (1999). "Cloning of Rab GTPases expressed in human skeletal muscle: studies in insulin-resistant subjects". Horm. Metab. Res. 30 (11): 656–62. doi:10.1055/s-2007-978953. PMID 9918381.
- Hartley JL, Temple GF, Brasch MA (2001). "DNA cloning using in vitro site-specific recombination". Genome Res. 10 (11): 1788–95. doi:10.1101/gr.143000. PMC 310948. PMID 11076863.
- Simpson JC, Wellenreuther R, Poustka A, et al. (2001). "Systematic subcellular localization of novel proteins identified by large-scale cDNA sequencing". EMBO Rep. 1 (3): 287–92. doi:10.1093/embo-reports/kvd058. PMC 1083732. PMID 11256614.
- Strausberg RL, Feingold EA, Grouse LH, et al. (2003). "Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences". Proc. Natl. Acad. Sci. U.S.A. 99 (26): 16899–903. doi:10.1073/pnas.242603899. PMC 139241. PMID 12477932.
- Gerhard DS, Wagner L, Feingold EA, et al. (2004). "The status, quality, and expansion of the NIH full-length cDNA project: the Mammalian Gene Collection (MGC)". Genome Res. 14 (10B): 2121–7. doi:10.1101/gr.2596504. PMC 528928. PMID 15489334.
- Wiemann S, Arlt D, Huber W, et al. (2004). "From ORFeome to biology: a functional genomics pipeline". Genome Res. 14 (10B): 2136–44. doi:10.1101/gr.2576704. PMC 528930. PMID 15489336.
- Mehrle A, Rosenfelder H, Schupp I, et al. (2006). "The LIFEdb database in 2006". Nucleic Acids Res. 34 (Database issue): D415–8. doi:10.1093/nar/gkj139. PMC 1347501. PMID 16381901.
This article is issued from Wikipedia. The text is licensed under Creative Commons - Attribution - Sharealike. Additional terms may apply for the media files.