Streptomyces sp. myrophorea
Streptomyces sp. myrophorea, isolate McG1 is a species of Streptomyces, that originates from a (ethnopharmacology) folk cure in the townland of Toneel North in Boho, County Fermanagh.[1] This area was previously occupied by the Druids (~1500 years ago) and before this neolithic people (~ 3,700 years ago) who engraved the nearby Reyfad stones.[2] Streptomyces sp. myrophorea is inhibitory to many species of ESKAPE pathogens, can grow at high pH (10.5) and can tolerate relatively high levels of radioactivity.[1]
| Streptomyces sp. myrophorea | |
|---|---|
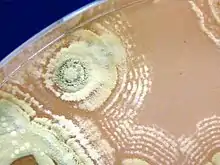 | |
| Mature Streptomyces sp myrophorea showing dusty green colonies with feint concentric rings | |
| Scientific classification | |
| Domain: | |
| Phylum: | |
| Order: | |
| Family: | |
| Genus: | |
| Species: | Streptomyces species |
| Variety: | Streptomyces sp. myrophorea isolate McG1 |
Physiology and morphology
Streptomyces sp. myrophorea isolate McG1 has light green to white spores and hyphae when cultivated on SFM agar.[1]

The colonies of Streptomyces sp. myrophorea have a distinctly dusty appearance and produce an aroma similar to germaline on maturation.[1] This bacteria produces many spores, approximately 0.5-1.0 micrometers in width which form in straight chains.[1]
Ecology
Streptomyces sp. myrophorea isolate McG1 was discovered in an alkaline, species rich environment.

This bacteria grows at a maximum pH of 10.5, and is therefore alkaliphilic. The bacteria tolerate higher levels of alkalinity but do not thrive.[1]
Streptomyces sp. myrophorea can also withstand relatively high levels of radiation (up to 4kGy. This may be related to the underlying limestone and shale substrata which emits radon gas.[1]
Antibiotic production
Only antibiotic gene synthesis clusters have been identified in Streptomyces sp. myrophorea; the antibiotics actually produced in-situ have yet to be identified.[1] Streptomyces sp. McG1 is broadly inhibitory to both Gram positive and Gram negative bacteria including carbapenem resistant Acinetobacter baumannii, (a critical pathogen on the World Health Organization priority pathogens list), vancomycin resistant Enterococcus faecium, methicillin resistant Staphylococcus aureus (listed as high priority) and Klebsiella pneumoniae.[1][3] Streptomyces sp. myrophorea has limited effects against strains of Enterococcus faecium and Pseudomonas aeruginosa.[1]
Alkaline tolerance
Streptomyces sp. myrophorea may be able to flourish in an alkaline environment because it contains many genes similar to those associated with alkaline tolerance in other bacterial species.[4][5]
Streptomyces sp. myrophorea list of alkaline tolerance genes:[1]
| Gene | Protein |
|---|---|
| 504 | Alkaline shock protein 23 |
| 6561 | Alkaline shock protein 23 |
| 1049 | Ammonium/H+ antiporter subunit amhM |
| 2800 | Aspartate/alanine antiporter |
| 2801 | Aspartate/alanine antiporter |
| 6218 | Aspartate/alanine antiporter |
| 3284 | Enhanced intracellular survival protein |
| 159 | K+/H+ antiporter nhaP |
| 3788 | K+/H+ antiporter nhaP2 |
| 1050 | K+/H+ antiporter yhaU |
| 2193 | K+/H+ antiporter yhaU |
| 1325 | K+ insensitive pyrophosphate- energized proton pump |
| 923 | Multidrug resistance protein mdtH |
| 3240 | Multidrug resistance protein mdtH |
| 3252 | Multidrug resistance protein mdtH |
| 6781 | Multidrug resistance protein mdtH |
| 1282 | Na+/H+ antiporter nhaA |
| 4210 | Na+/H+ antiporter subunit A |
| 4211 | Na+/H+ antiporter subunit C |
| 4212 | Na+/H+ antiporter subunit D |
| 4215 | Na+/H+ antiporter subunit G |
| 660 | Na+/(H+or K+) antiporter gerN |
| 3425 | Na+/(H+or K+) antiporter gerN |
| 7210 | putative Na+/H+ exchanger |
| 1727 | Sodium, potassium, lithium and rubidium/H+ antiporter |
In-vitro antibiotic resistance
The presence of antibiotic resistance genes is often linked to the production of antibiotics.[6] Stretpomyces sp. myrophorea has been recorded to be resistant to 28 antibiotics and sensitive to eight antibiotics in one series of tests. The antibiotics were tested at breakpoint concentrations recommended by The European Committee on Antimicrobial Susceptibility Testing - EUCAST, to test antimicrobial susceptibility.[1][7]
Antibiotic sensitivity of Streptomyces sp. myrophorea:[1] Sensitivities recorded as: Sensitivite (S), Resistant (R) or Intermediate (I).
| Antibiotic | Quantity (µg) | Sensitivity |
|---|---|---|
| Amikacin | 30 | S |
| Ampicillin | 30 | R |
| Ampicillin + sulbactam | 20 | R |
| Augmentin | 30 | R |
| Apromycin | 20 | R |
| Carbenicillin | 20 | R |
| Cefepime | 30 | R |
| Ceftazidime | 10 | R |
| Ceftibuten | 30 | R |
| Cefoxitin | 30 | R |
| Ceftriaxone | 30 | R |
| Cefuroxime | 30 | R |
| Cefurin | 5 | R |
| Ciprofloxacin | 5 | R |
| Clavuranic acid | 10 | R |
| Ertapenem | 10 | R |
| Erythromycin | 15 | I |
| Gentamicin | 10 | S |
| Imipenem | 10 | R |
| Kanamycin | 20 | R |
| Linezolid | 10 | I |
| Meropenem | 10 | R |
| Mupiricin | 200 | R |
| Netilmicin | 10 | S |
| Nitrofurantoin | 100 | R |
| Novobiocin | 5 | R |
| Piperacillin + tazobactam | 36 | R |
| Quinpristin + dalropristin | 5 | R |
| Rifampicin | 5 | R |
| Streptomycin | 20 | R |
| Tigecycline | 15 | R |
| Tetracycline | 20 | R |
| Trimethoprim + sulfamethoxazole | 25 | R |
| Vancomycin | 5 | R |
Whole genome sequencing
The genome sequence of Streptomyces sp. myrophorea, isolate McG1 was deposited at the NCBI (TaxID 2099643), Biosample accession number SAMN08518548, BioProject accession number PRJNA433829, Submission ID: SUB3653175 Locus tag prefix: C4625. https://www.ncbi.nlm.nih.gov/bioproject/433829.[1] Link to NCBI sequence read archive (SRA) https://www.ncbi.nlm.nih.gov/sra?LinkName=biosample_sra&from_uid=8518548 Copies of this bacteria are deposited in the National Collection of Type Cultures (NCTC), UK[8] and the Deutsche Sammlung von Mikroorganismen und Zellkulturen (DSMZ) GmbH, Germany.[9]
References
- Terra, L.; et al. (2018). "A Novel Alkaliphilic Streptomyces Inhibits ESKAPE Pathogens". Frontiers in Microbiology. 9: 2458. doi:10.3389/fmicb.2018.02458. PMC 6232825. PMID 30459722.
- Andrew Halpin; Conor Newman (2006). Ireland: an Oxford archaeological guide to sites from earliest times to AD 1600. Oxford University Press. p. 8. ISBN 978-0-19-280671-0.
- "Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics". World Health Organization.
- Holdsworth, S. R.; Law, C. J. (2013). "Multidrug Resistance Protein MdtM Adds to the Repertoire of Antiporters Involved in Alkaline pH Homeostasis in Escherichia coli". BMC Microbiology. 13: 113. doi:10.1186/1471-2180-13-113. PMC 3668916. PMID 23701827.
- Krulwich, T. A.; Sachs, G.; Padan, E. (2011). "Molecular Aspects of Bacterial pH Sensing and Homeostasis". Nature Reviews Microbiology. 9 (5): 330–343. doi:10.1038/nrmicro2549. PMC 3247762. PMID 21464825.
- Nodwell, J.R. (2007). "Novel Links between Antibiotic Resistance and Antibiotic Production". Journal of Bacteriology. 189 (10): 3683–3685. doi:10.1128/JB.00356-07. PMC 1913332. PMID 17384190.
- "The European Committee on Antimicrobial Susceptibility Testing - EUCAST". Retrieved 2018-10-13.
- "National Collection of Type Cultures (NCTC)". Public Health England. Retrieved 2018-10-13.
- "Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures". Leibniz-Institut DSMZ-Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH. Retrieved 2018-10-13.
External links
- Terra, Luciana; Dyson, Paul J.; Hitchings, Matthew D.; Thomas, Liam; Abdelhameed, Alyaa; Banat, Ibrahim M.; Gazze, Salvatore A.; Vujaklija, Dušica; Facey, Paul D.; Francis, Lewis W.; Quinn, Gerry A. (2018). "A Novel Alkaliphilic Streptomyces Inhibits ESKAPE Pathogens". Frontiers in Microbiology. 9: 2458. doi:10.3389/fmicb.2018.02458. PMC 6232825. PMID 30459722.
- "Streptomyces McG#1 from Boho in Northern Ireland".
- Terra, Luciana; Dyson, Paul J.; Hitchings, Matthew D.; Thomas, Liam; Abdelhameed, Alyaa; Banat, Ibrahim M.; Gazze, Salvatore A.; Vujaklija, Dušica; Facey, Paul D.; Francis, Lewis W.; Quinn, Gerry A. (2018). "A Novel Alkaliphilic Streptomyces Inhibits ESKAPE Pathogens". Frontiers in Microbiology. 9: 2458. doi:10.3389/fmicb.2018.02458. PMC 6232825. PMID 30459722.