Balancer chromosome
Balancer chromosomes (or balancers) are a genetic tool for maintenance of recessive lethal (or sterile) mutations without active selection. Since such mutations have to be maintained as heterozygotes, they continually lead to production of wild-type organisms, which can be prevented by replacing the homologous wild-type chromosome with a balancer. In that capacity, balancers are crucial for research on the model organism Drosophila melanogaster (fruit fly), whose stocks cannot be archived (e.g. frozen). They can also be used in forward genetic screens to specifically identify recessive lethal (or sterile) mutations. For that reason, balancers are also used in other model organisms, most notably the worm C.elegans and the mouse.[1]
Typical balancer chromosomes:
- carry recessive lethal mutations themselves, eliminating homozygotes which do not carry the desired mutation
- suppress meiotic recombination with their homologs, which prevents de novo creation of wild-type chromosomes
- carry dominant markers, which can help identify rare recombinants and which are useful for screening purposes
History
Balancer chromosomes were first used in the fruit fly by Hermann Muller, the pioneer of using radiation for organismal mutagenesis.[2]
In the modern usage of balancer chromosomes, random mutations are first caused, usually by feeding the larva ethyl methane sulphonate (EMS). When a phenotype of interest is observed, the line is crossed with another line containing balancer chromosomes to maintain their lineage.[3] In one instance they were used to genetically screen a population of Caenorhabditis elegans. At this point in time scientists had already realized the benefits of being able to genetically screen populations of organisms for genetic study. Equally as important, they also realized that they could limit crossing over in these populations as well give them a very consistent genetic population.[4]
The use of balancer chromosomes has evolved into a well known and widely used method for genetic screening of model organisms. They are even being used to investigate the role of heterochromatin packing and the effect it has on genes[5] as well as studies on the effect telomeres have on gene silencing.[6]
Mechanism
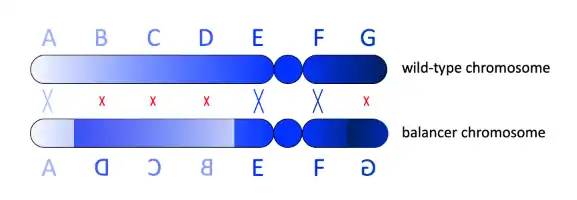
Mutations without recessive lethal (or sterile) phenotype in diploid organisms can simply be bred to homozygosity and maintained by crossing homozygotes. However, homozygotes for recessive lethal mutations are, per definition, non-viable, which necessitates maintenance of the mutation in heterozygous organisms. Crosses between heterozygotes yield a wild-type organisms, in addition to hetereozygotes and the non-viable homozygotes. To maintain a purely heterozygous line, wild-type offspring needs to be identified and prevented from mating. This can be prohibitively resource intensive, especially if long-term maintanance of the mutation is the goal.
Substituting a balancer chromosome for the wild-type chromosome homologous to the chromosome carrying the mutation prevents the establishment of wild-type organisms in various ways. First, a balancer carries a recessive lethal mutation, which makes the organism non-viable if two copies of balancer are inherited (ie no copy of the desired mutation). However, recombination between the balancer and the mutated allele may result in a creation of a wild-type chromosome. To suppress recombination, balancers usually harbor multiple, nested chromosomal inversions so that synapsis between homologous chromosomes is disrupted.[7] If crossing over does occur, it is often unbalanced, with each resulting chromatid lacking some genes and carrying two copies of others. It can also lead to dicentric or acentric chromosomes (chromosomes with two centromeres or no centromere), which end up breaking up and mutating or being lost. All of these outcomes are very likely to be lethal. Finally, balancer chromosomes carry dominant markers such as genes for green fluorescent protein or enzymes that make pigments, which allow researchers to easily recognize organisms that carry the balancer.[8] In the unlikely case of viable recombination, the marker may be lost thus alerting the researchers of the event.
Importantly, suppression of recombination by nested inversions only occurs at the inverted intervals, while other regions (usually peri-centromeric and sub-telomeric) are free to recombine. Likewise, if the desired mutation is in the same locus as the balancer's recessive lethal mutation (i.e. is in strong linkage disequilibrium with it), recombination resulting in a wild-type chromosome is very unlikely, regardless of recombination suppressive inversions.
In addition to simply maintaining an isolated recessive lethal (or sterile) mutation, balancer chromosomes are also used in forward genetic screens to identify such mutations. In such screens randomly mutagenized organisms carrying a balancer are crossed with each other. Offspring that carries the balancer, identified by the dominant marker, can be crossed with littermates. Any such cross that does not produce marker-negative animals is likely the result of a recessive lethal mutation in the non-balancer chromosome. Of course, only the genomic interval covered by the inversions in the balancer can be screened in this way, with recessive lethal mutations in other intervals and on other chromosomes being lost.
Naming convention in Drosophila
Balancer chromosomes are named for the chromosome they serve to stabilize and for the phenotypic or genetic marker the balancer carries.[9] The naming of balancer chromosomes in D. melanogaster has been standardized as follows: the first letter of the chromosome's name represents the number of the chromosome it stabilizes. F stands for the first chromosome, S stands for second, and T stands for third. The small fourth chromosome does not undergo recombination and therefore does not require balancing. This letter is then followed by an M, for "multiply inverted." The M is followed by a number to distinguish balancers of the same chromosome. Additionally, the genetic marker or markers of the balancer are listed after the name and separated by a comma. Generally mutations with easily observable dominant phenotypic traits that are often homozygous lethal are used to ensure that all progeny are heterozygous. For example, the commonly used "TM3, Sb" balancer is a balancer chromosome that stabilizes the third chromosome and carries a mutant Sb ("stubble") gene as a marker. All flies containing the TM3, Sb balancer will have shortened (or stubbly) hairs on the back of the fly, which are easily seen when viewed through a microscope. The 3 is to distinguish this from other third chromosome balancers, such as "TM1" and "TM2".
A line is said to be "double-balanced" if it is heterozygous for two different balancer chromosomes (for example, TM6,Tb/TM3,Ser) on a balancer chromosome and a homozygous lethal, heterozygous visible mutant on the other, wild-type chromosome (for example, D/TM3,Ser). Most balancer chromosomes also carry a recessive allele such as the "ebony" mutation that is only manifest in these stocks with two balancer chromosomes. These stocks are often used to provide sources of easily traceable traits when breeding two different lines together so that the correct progeny of each cross might be selected. Stocks double-balanced at both the second and third chromosomes in Drosophila are widely available.
Important scientific contributions using balancer chromosomes
Balancer chromosomes already give geneticists a reliable method for genetically screening organisms for a mutation and keeping that line constant. A new technique using balancer chromosomes is explored in the paper "The Autosomal Flp-Dfs Technique for Generating Germline Mosaics in Drosophila Melanogaster." This paper showed for the first time that it is possible to screen for a recessive mutation that only shows phenotype when homozygous. Using old balancer chromosome methods, genetic screening only allowed the picking out of heterozygous dominant mutations. This experiment uses clonal screening to detect homozygous individuals and keep them in a constant line.[10]
They achieved this by using a gene isolated from yeast. This gene is called FLP recombinase and causes large chromosomal inversions. Through trial and error they found that the chromosomes could be recombined such that each had the recessive mutation while the other half contained half of a balancer chromosome with a physical marker and a lethal recessive. The other homolog did not contain the lethal recessive in the lines that survived. Figure one in the paper illustrates the screen. This new technique allowed recessive screening in 95% of the Drosophila genome. It also greatly improved yields in germ line mutations.[10]
Another published paper that employed the use of balancer chromosomes is "Inhibition of RNA Interference and Modulation of Transposable Element Expression by Cell Death in Drosophila." This paper demonstrates the power of balancer chromosomes and what can be accomplished with genetically stable lines. A line was established that exhibited low levels of cell death and was named EGFPir hs-hid. The RNAi levels were analyzed and they found interesting results in the cells undergoing low levels of cell death and the surrounding cells in the tissue. They found that these cells would shut down their RNAi mechanism via maintaining RNA in a double stranded state. If RNA remains in a double stranded state then the RNAi mechanism of gene silencing is shut down.
The authors speculated that this response was an evolutionary trend toward redundant immune response against RNA viruses. If one cell is already undergoing cell death to attempt to stop spread of a virus, then the RNAi immune response has been ineffective. This causes another immune response that attempts to stop the virus which is binding double stranded RNA and keeping double stranded so that it cannot be transcribed into virus proteins. The mechanism of maintaining double stranded RNA is not known.[11]
References
- Zheng, Binhai; Marijke Sage; Wei-Wen Cai; Debrah M. Thompson; Beril C. Tavsanli; Yin-Chai Cheah; Allan Bradley (1998). "Engineering a mouse balancer chromosome". Nature Genetics. 22 (4): 375–378. doi:10.1038/11949. PMID 10431243.
- Hermann Muller Invented the Balancer Chromosome
- Lewis, E. B.; F. Bacher (1968). "Methods of feeding ethyl methane sulphonate (EMS) to Drosophila males". Drosophila Information Service. 43: 193.
- Herman, Robert K.; Albertson, Donna G.; Brenner, Sydney (1976-05-15). "Chromosome Rearrangements in Caenorhabditis Elegans". Genetics. 83 (1): 91–105. ISSN 0016-6731. PMC 1213508. PMID 1269921. Retrieved 2015-05-11.
- Bushy, Daniel; John Locke (November 1, 2004). "Mutations in Su(var)205 and Su(var)3-7 Suppress P-Element-Dependent Silencing in Drosophila melanogaster". Genetics. 3. 168 (3): 1395–1411. doi:10.1534/genetics.104.026914. PMC 1448784. PMID 15579693.
- Mason, James; Random Joshua; Konev Alexander (November 1, 2004). "A Deficiency Screen for Dominant Suppressors of Telomeric Silencing in Drosophila". Genetics. 3. 168 (3): 1353–1370. doi:10.1534/genetics.104.030676. PMC 1448782. PMID 15579690.
- Kile, Benjamin T.; Kathryn E. Hentges; Amander T. Clark; Hisashi Nakamura; Andrew P. Salinger; Bin Liu; Neil Box; David W. Stockton; Randy L. Johnson; Richard R. Behringer; Allan Bradley; Monica J. Justice (4 September 2003). "Functional genetic analysis of mouse chromosome 11". Nature. 425 (6953): 81–86. doi:10.1038/nature01865. PMID 12955145.
- Casso, David; Felipe-Andrés Ramírez-Weber; Thomas B. Kornberg (March 2000). "GFP-tagged balancer chromosomes for Drosophila melanogaster". Mechanisms of Development. 91 (1–2): 451–454. doi:10.1016/S0925-4773(00)00248-3. PMID 10704882.
- Fly Pushing: The Theory and Practice of Drosophila Genetics By Ralph J. Greenspan. Page 13
- Chou, T. B.; N. Perrimon (December 1996). "The Autosomal Flp-Dfs Technique for Generating Germline Mosaics in Drosophila Melanogaster". Genetics. 144 (4): 1673–1679. PMC 1207718. PMID 8978054.
- Xie, Weiwu; Liang Chengzhi; James Birchler (1 August 2011). "Inhibition of RNA Interference and Modulation of Transposable Element Expression by Cell Death in Drosophila". Genetics. 188 (4): 823–834. doi:10.1534/genetics.111.128470. PMC 3176087. PMID 21596898. Retrieved 2011-11-22.
