Carmovirus
Carmovirus is a genus of viruses, in the family Tombusviridae. Plants serve as natural hosts. There are currently 19 species in this genus including the type species Carnation mottle virus.[1][2] It is classified under the Baltimore classification system as a group IV virus, having a single-stranded, positive-sense RNA genome.[3]
| Carmovirus | |
|---|---|
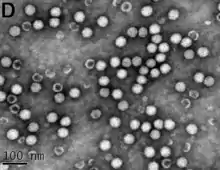 | |
| Transmission electron micrograph of bean mild mosaic virus (BMMV) virions | |
| Virus classification | |
| Group: | Group IV ((+)ssRNA) |
| Family: | |
| Genus: | Carmovirus |
| Type species | |
History
Proposed at 1987 ICTV meeting as an unassigned genus and assigned to Tombusviridae in 1993,[4] the ICTV recognises 19 species, as of the 2013 release.[3][5]
At the 1987 meeting the following were proposed as members: Carnation mottle virus, Turnip crinkle virus, Galinsoga mosaic virus, Glycine mottle virus, Hibiscus chlorotic ringspot virus (HCRSV), Pelargonium flower break virus, Saguaro cactus virus. Galinsoga mosaic virus and Glycine mottle virus are not currently members.
In addition the following were proposed as tentative members: Bean mild mosaic virus, Blackgram mottle virus, *Cowpea mottle virus, Elderberry latent virus, *Melon necrotic spot virus, Narcissus tip necrosis virus, Plantain 6 virus, Tephrosia symptomless virus. (Taxonomic proposal 30) Only those marked with an *asterisk have been accepted up to the 2013 release.
Taxonomy
Group: ssRNA(+)
- Family: Tombusviridae
- Genus: Carmovirus
- Ahlum waterborne virus
- Angelonia flower break virus
- Bean mild mosaic virus
- Calibrachoa mottle virus
- Cardamine chlorotic fleck virus
- Carnation mottle virus
- Cowpea mottle virus
- Cucumber soil-borne virus
- Hibiscus chlorotic ringspot virus
- Honeysuckle ringspot virus
- Japanese iris necrotic ring virus
- Melon necrotic spot virus
- Nootka lupine vein clearing virus
- Pea stem necrosis virus
- Pelargonium flower break virus
- Saguaro cactus virus
- Soybean yellow mottle mosaic virus
- Turnip crinkle virus
- Weddel waterborne virus
Structure
Viruses in Carmovirus are non-enveloped, with icosahedral and spherical geometries, and T=3 symmetry. The diameter is around 28-34 nm, made up of 32 capsomers. Genomes are linear, around 4-5.4kb in length. Each subunit consists of 180 proteins. By weight, the viron is 80% protein and 20% nucleic acids.[1]
| Genus | Structure | Symmetry | Capsid | Genomic arrangement | Genomic segmentation |
|---|---|---|---|---|---|
| Carmovirus | Icosahedral | T=3 | Non-enveloped | Linear | Monopartite |
Life cycle
Viral replication is cytoplasmic. Entry into the host cell is achieved by penetration into the host cell. Replication follows the positive stranded RNA virus replication model. Positive stranded rna virus transcription, using the premature termination model of subgenomic rna transcription is the method of transcription. Translation takes place by viral initiation, and suppression of termination. The virus exits the host cell by tubule-guided viral movement. Plants serve as the natural host. Transmission routes are mechanical, seed borne, and contact.[1]
| Genus | Host details | Tissue tropism | Entry details | Release details | Replication site | Assembly site | Transmission |
|---|---|---|---|---|---|---|---|
| Carmovirus | Plants | None | Viral movement; mechanical inoculation | Viral movement | Cytoplasm | Cytoplasm | Mechanical: contact; seed |
Recombination
RNA recombination occurs frequently during replication of carmoviruses, facilitating viral genome repair and promoting sequence variability.[6] Recombination occurs by a mechanism in which the viral RNA-dependent RNA polymerase switches templates during RNA synthesis (copy choice recombination).[6]
Epidemiology
The virus' natural host is a species of Caryophyllaceae, responsible for carnation mottle, but it can infect a far larger range of plants. Symptoms are localised lesions where cell death has occurred. The virus spreads via arthropod vectors carrying infected sap. Although the disease is widespread in carnations, the symptoms are relatively mild and do not pose a major problem. In other plants however, such as the Calla lily, the disease is of more concern, as the plant is of significant economic importance to Taiwan.
References
- "Viral Zone". ExPASy. Retrieved 15 June 2015.
- ICTV. "Virus Taxonomy: 2014 Release". Retrieved 15 June 2015.
- Unknown author (2004). 00.074.0.02. Carmovirus. In: ICTVdB - The Universal Virus Database, version 3. Büchen-Osmond, C. (Ed), ICTVdB Management, Columbia University, New York, USA
- "MINUTES OF THE 7th MEETING OF THE ICTV EDMONTON, CANADA, 12th AUGUST 1987" (PDF). Archived from the original (PDF) on 13 December 2014. Retrieved 11 December 2014.
- ICTV Taxonomy History for Carmovirus
- Cheng CP, Nagy PD. Mechanism of RNA recombination in carmo- and tombusviruses: evidence for template switching by the RNA-dependent RNA polymerase in vitro. J Virol. 2003 Nov;77(22):12033-47. doi: 10.1128/jvi.77.22.12033-12047.2003. PMID: 14581540; PMCID: PMC254248
