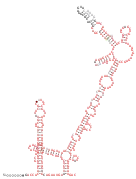
Coronavirus 3′ UTR
Coronavirus genomes are positive-sense single-stranded RNA molecules with an untranslated region (UTR) at the 3′ end which is called the 3′ UTR. The 3′ UTR is responsible for important biological functions, such as viral replication.[1] The 3′ UTR has a conserved RNA secondary structure but different Coronavirus genera (Alpha-, Beta-, Gamma-, and Deltacoronaviruses) have different structural features described below.
Alphacoronavirus 3′ UTR

The 3′ UTR of Alphacoronaviruses consists of two small hairpins (PK-SL2) which can form an alternate conformation (PK-SL1) where the loop region of the second hairpin interacts with the stem of the first hairpin (see Coronavirus 3′ UTR pseudoknot). PK-SL2 has been confirmed in HCoV-229E and HCoV-NL63 by in vitro structure probing experiments. Downstream of this pseudoknot lies the hypervariable region (HVR), which is supported by many covarying base pairs in alphacoronaviruses. Further, a conserved octanucleotide sequence 5′-GGAAGAGC-3′ is present in the HVR.[2]
Betacoronavirus 3′ UTR
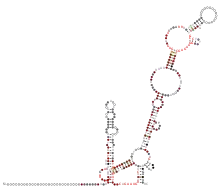
The 3′ UTR of Betacoronaviruses is approximately 300–500 nucleotides long. The most 5′ structure is the bulged stem-loop (BSL), starting downstream of the N gene stop codon and is essential for viral replication. It is predicted to be conserved among Betacoronaviruses. Downstream to the BSL lies the hypervariable bulged-stemloop region (HVR), containing a triple helix junction. The hairpin stem-loop arising from this junction participates in the pseudoknot pairing with the BSL region. The pseudoknot and the BSL partially overlap and therefore cannot be formed at the same time. It is proposed that the BSL and pseudoknot act as a molecular switch that may regulate a transition during viral RNA synthesis. Part of the HVR is the octanucleotide sequence 5′-GGAAGAGC-3′ which is conserved among Alpha- and Betacoronaviruses. The 3′ UTR of SARS-CoV-2 is similar to other Betacoronaviruses and is approximately 300–500 nucleotides long.
Gammacoronavirus 3′ UTR
In Gammacoronaviruses a stem-loop located at the 3′ UTR is observed, which is vital for viral replication. Even though a nearby pseudoknot structure, which has been observed in other genera of coronaviruses, is present, its functional importance has not been established yet.
Deltacoronavirus 3′ UTR
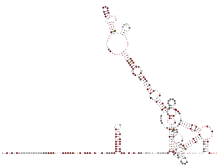
There are two smaller hairpins and the hypervariable region (HVR) present in the 3′ UTR of Deltacoronaviruses. The pseudoknot structure which is typical for Alpha- and Betacoronaviruses has not been predicted in Deltacoronaviruses so far.
See also
References
- Madhugiri, R.; Fricke, M.; Marz, M.; Ziebuhr, J. (2016). "Coronavirus cis-Acting RNA Elements". Advances in Virus Research. 96: 127–163. doi:10.1016/bs.aivir.2016.08.007. ISBN 978-0-12-804736-1. ISSN 1557-8399. PMC 7112319. PMID 27712622.
- Yang, Dong; Leibowitz, Julian L. (2015-08-03). "The structure and functions of coronavirus genomic 3′ and 5′ ends". Virus Research. 206: 120–133. doi:10.1016/j.virusres.2015.02.025. ISSN 1872-7492. PMC 4476908. PMID 25736566.