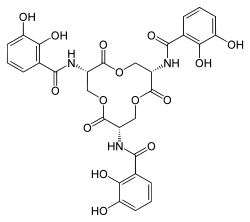
Enterobactin
Enterobactin (also known as enterochelin) is a high affinity siderophore that acquires iron for microbial systems. It is primarily found in Gram-negative bacteria, such as Escherichia coli and Salmonella typhimurium.[1]
 | |
| Names | |
|---|---|
| IUPAC name
N,N',N''-((3S,7S,11S)-2,6,10- trioxo-1,5,9-trioxacyclododecane- 3,7,11-triyl)tris(2,3-dihydroxybenzamide) | |
| Identifiers | |
3D model (JSmol) |
|
| ChEBI | |
| ChEMBL | |
| ChemSpider | |
PubChem CID |
|
| UNII | |
CompTox Dashboard (EPA) |
|
| |
| |
| Properties | |
| C30H27N3O15 | |
| Molar mass | 669.55 g/mol |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa). | |
| Infobox references | |
Enterobactin is the strongest siderophore known, binding to the ferric ion (Fe3+) with the affinity (K = 1052 M−1).[2] This value is substantially larger than even some synthetic metal chelators, such as EDTA (Kf,Fe3+ ~ 1025 M−1).[3] Due to its high affinity, enterobactin is capable of chelating even in environments where the concentration of ferric ion is held very low, such as within living organisms. Enterobactin can extract iron even from the air.[4] Pathogenic bacteria can steal iron from other living organisms using this mechanism, even though the concentration of iron is kept extremely low due to the toxicity of free iron.
Structure and biosynthesis
Chorismic acid, an aromatic amino acid precursor, is converted to 2,3-dihydroxybenzoic acid (DHB) by a series of enzymes, EntA, EntB and EntC. An amide linkage of DHB to L-serine is then catalyzed by EntD, EntE, EntF and EntB. Three molecules of the DHB-Ser formed undergo intermolecular cyclization, yielding enterobactin.[5] Although a number of stereoisomers are possible due to the chirality of the serine residues, only the Δ-cis isomer is metabolically active.[3] The first three-dimensional structure of a metal enterobactin complex was determined as the vanadium(IV) complex.[6] Although ferric enterobactin long eluded crystallization, its definitive three-dimensional structure was ultimately obtained using racemic crystallography, in which crystals of a 1:1 mixture of ferric enterobactin and its mirror image (ferric enantioenterobactin) were grown and analyzed by X-ray crystallography.[7]

Mechanism
Iron deficiency in bacterial cells triggers secretion of enterobactin into the extracellular environment, causing formation of a coordination complex "FeEnt" wherein ferric ion is chelated to the conjugate base of enterobactin. In Escherichia coli, FepA in the bacterial outer membrane then allows entrance of FeEnt to the bacterial periplasm. FepB,C,D and G all participate in transport of the FeEnt through the inner membrane by means of an ATP-binding cassette transporter.[5]
Due to the extreme iron binding affinity of enterobactin, it is necessary to cleave FeEnt with ferrienterobactin esterase to remove the iron. This degradation yields three 2,3-dihydroxybenzoyl-L-serine units. Reduction of the iron (Fe3+ to Fe2+) occurs in conjunction with this cleavage, but no FeEnt bacterial reductase enzyme has been identified, and the mechanism for this process is still unclear.[8] The reduction potential for Fe3+/Fe2+–enterobactin complex is pH dependent and varies from −0.57 V (vs NHE) at pH 6 to −0.79 V at pH 7.4 to −0.99 at pH values higher than 10.4.[9]
History
Enterobactin was discovered by Gibson and Neilands groups in 1970.[10][11] These initial studies established the structure and its relationship to 2,3-dihydroxybenzoic acid.
References
- Dertz EA, Xu J, Stintzi A, Raymond KN (January 2006). "Bacillibactin-mediated iron transport in Bacillus subtilis". Journal of the American Chemical Society. 128 (1): 22–3. doi:10.1021/ja055898c. PMID 16390102.
- Carrano CJ, Raymond KN (1979). "Ferric Ion Sequestering Agents. 2. Kinetics and Mechanism of Iron Removal From Transferrin by Enterobactin and Synthetic Tricatechols". J. Am. Chem. Soc. 101 (18): 5401–5404. doi:10.1021/ja00512a047.
- Walsh CT, Liu J, Rusnak F, Sakaitani M (1990). "Molecular Studies on Enzymes in Chorismate Metabolism and the Enterobactin Biosynthetic Pathway". Chemical Reviews. 90 (7): 1105–1129. doi:10.1021/cr00105a003.
- "Enterobactin". CLOUD-CLONE CORP.
- Raymond KN, Dertz EA, Kim SS (April 2003). "Enterobactin: an archetype for microbial iron transport". Proceedings of the National Academy of Sciences of the United States of America. 100 (7): 3584–8. doi:10.1073/pnas.0630018100. PMC 152965. PMID 12655062.
- Karpishin TB, Raymond KN (1992). "The First Structural Characterization of A Metal-Enterobactin Complex: [V(enterobactin)]2-". Angewandte Chemie International Edition in English. 31 (4): 466–468. doi:10.1002/anie.199204661.
- Johnstone TC, Nolan EM (October 2017). "Determination of the Molecular Structures of Ferric Enterobactin and Ferric Enantioenterobactin Using Racemic Crystallography". Journal of the American Chemical Society. 139 (42): 15245–15250. doi:10.1021/jacs.7b09375. PMC 5748154. PMID 28956921.
- Ward TR, Lutz A, Parel SP, Ensling J, Gütlich P, Buglyó P, Orvig C (November 1999). "An Iron-Based Molecular Redox Switch as a Model for Iron Release from Enterobactin via the Salicylate Binding Mode". Inorganic Chemistry. 38 (22): 5007–5017. doi:10.1021/ic990225e. PMID 11671244.
- Lee CW, Ecker DJ, Raymond KN (1985). "Coordination chemistry of microbial iron transport compounds. 34. The pH-dependent reduction of ferric enterobactin probed by electrochemical methods and its implications for microbial iron transport". J. Am. Chem. Soc. 107 (24): 6920–6923. doi:10.1021/ja00310a030.
- Pollack JR, Neilands JB (March 1970). "Enterobactin, an iron transport compound from Salmonella typhimurium". Biochemical and Biophysical Research Communications. 38 (5): 989–92. doi:10.1016/0006-291X(70)90819-3. PMID 4908541.
- O'Brien IG, Cox GB, Gibson F (March 1970). "Biologically active compounds containing 2,3-dihydroxybenzoic acid and serine formed by Escherichia coli". Biochimica et Biophysica Acta (BBA) - General Subjects. 201 (3): 453–60. doi:10.1016/0304-4165(70)90165-0. PMID 4908639.
