FAM63A
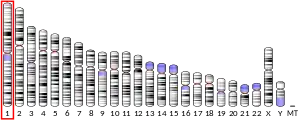
Family with sequence similarity 63, member A is a protein that, in humans, is encoded by the FAM63A gene. It is located on the minus strand of chromosome 1 at locus 1q21.3.[5]
Evolutionarily, FAM63A orthologs are found in most vertebrates, and distant homologs of FAM63A are found in invertebrates.[6] FAM63A is ubiquitously expressed throughout human tissues, and it is present during every stage of development.[7]
It has been linked to a biomarker in Chronic Kidney Disease and Alzheimer's Disease.[8]
Gene
Locus
FAM63A is located on the minus strand of chromosome 1 at band 1q21.3, spanning 11,829 bp. Other genes surrounding FAM63A include ANXA9 and Prune.[9]
Aliases
FAM63A has two aliases KIAA1390 and PR11-316M1.5.[10]
mRNA
Primary structure
In humans, there are four isoforms of FAM63A, and there are 10 predicted isoforms. Isoform 1 of FAM63A has a molecular weight of 51.8 kilodaltons, and it contains 11 exons.[11][12] The different isoforms tend to differ at the 5' or 3' end by truncation. Transcription produces 23 introns, 14 spliced variants, and 6 unspliced forms.[13]
Protein

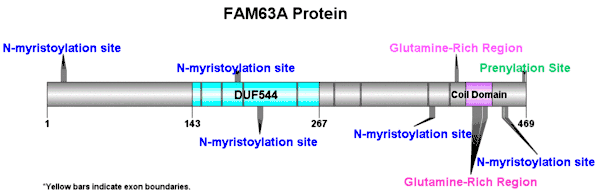
Domains and motifs
FAM63A contains a domain of unknown function (DUF 544). DUF544 contains 125 amino acids, running from Met143 to Thr267.[14] Although not completely conserved, this domain is highly conserved across vertebrates, invertebrates, and plants.[15] FAM63A does not contain a transmembrane domain, and it is found primarily in nuclear regions of the cell. [16]
Two repeats of four glutamines are seen from amino acid 400-403 and from amino acid 426-429, leading to an elevated glutamine composition at the C-terminus.
Composition
FAM63A is composed of 469 amino acids.[17] There is an increased presence of glutamine found near the C terminus making FAM63A glutamine rich. FAM63A contains a greater amount of negatively charged (acidic) amino acids than positively charged (basic) amino acids which makes FAM63A a slightly acidic protein. Acidic amino acids such as aspartic acid and glutamic acid are more prevalent than the basic amino acids such as lysine and arginine. This overall acidic composition gives FAM63A an acidic isoelectric point of 4.6.[18]
Post-translational modifications
FAM63A contains 25 phosphorylation sites in humans, including 12 serine, 10 threonine, and 3 tyrosine. Additionally, there are 5 N-myristoylation sites, and there is 1 prenylation site. FAM63A contains no glycosylation sites, transmembrane domains, or signal peptides. [19]
Secondary structure
The secondary structure for FAM63A has not been explicitly determined. There are, however, predictions for a possible secondary structure. There is a coiled-coil domain at the end of the protein, and in the predicted secondary structure, there is an alpha helix between amino acids 410 and 436. This helix is conserved throughout more distant orthologs of FAM63A. These data support each other, and it gives a confident prediction of the secondary structure.[20]
Homology/evolution
In FAM63A, there are several amino acids that are conserved in all vertebrates for which sequences are available. Gly239 is the only amino acid that is conserved in all vertebrates, invertebrates, and plants for which sequences are available. Because there is only one amino acid that is absolutely conserved, a possible function for the conserved Glycine was not deduced. The 25 amino acid sequence ranging from Val313 to Gly338 is the most highly conserved in all vertebrates, invertebrates, and plants for which sequences are available. Although the sequence is not absolutely conserved, it is very highly conserved, even in the most distantly related organisms like fungi and plants.
Orthologs
The protein FAM63A has several strict orthologs. These strict orthologs are found in organisms ranging from Primates to Fish.[23]
| Scientific Name | Common Name | Divergence | Accession Number | Length | Identity | Similarity | Query Cover |
|---|---|---|---|---|---|---|---|
| Homo sapiens | Humans | N/A | NP_060849.2 | 469 aa | 100% | 100% | 100% |
| Pan paniscus | Bonobo | 6.1 MYA | XP_003817322.1 | 469 aa | 99.1% | 99.4% | 100% |
| Mus musculus | House Mouse | 91.0 MYA | NP_955769.1 | 468 aa | 86.0% | 90.2% | 100% |
| Bos taurus | Cow | 97.4 MYA | NP_001039389.1 | 469 aa | 85.6% | 88.5% | 100% |
| Trichechus manatus latirostris | West Indian Manatee | 104.7 MYA | XP_004389621.1 | 465 aa | 84.9% | 89.8% | 100% |
| Sarcophilus harrisii | Tasmanian Devil | 176.1 MYA | XP_003769968.1 | 464 aa | 78.3% | 82.9% | 100% |
| Taeniopygia guttata | Zebra Finch | 324.0 MYA | XP_002191502.2 | 335 aa | 43.2% | 80.1% | 53% |
| Gallus gallus | Chicken | 324.5 MYA | XP_003642724 | 462 aa | 66.9% | 76.1% | 92% |
| Chrysemys picta bellii | Painted Turtle | 324.5 MYA | XP_005293753.1 | 525 aa | 62.0% | 87.2% | 78% |
| Pelodiscus sinensis | Chinese Softshell Turtle | 324.5 MYA | XP_006119467.1 | 502 aa | 61.6% | 77.4% | 94% |
| Alligator mississippiensis | American Alligator | 324.5 MYA | XP_006274676.1 | 520 aa | 59.90% | 77.50% | 86% |
| Pseudopodoces humilis | Ground Tit | 324.5 MYA | XP_005533539.1 | 502 aa | 58.0% | 82.3% | 78% |
| Anas platyrhynchos | Mallard | 324.5 MYA | XP_005026841.1 | 415 aa | 57.9% | 78.6% | 83% |
| Xenopus tropicalis | Western Clawed Frog | 361.2 MYA | XP_002937311.1 | 506 aa | 61.3% | 83.7% | 76% |
| Latimeria chalumnae | West Indian Ocean Coelacanth | 430.0 MYA | XP_006006147.1 | 513 aa | 44.7% | 86.9% | 55% |
| Danio rerio | Zebrafish | 454.6 MYA | XP_005159508.1 | 520 aa | 52.2% | 80.2% | 76% |
FAM63A evolved through time at a relatively moderate rate.
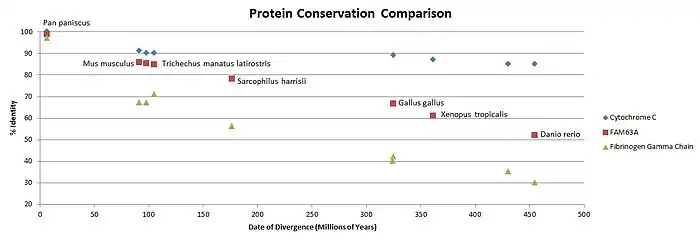
Paralogs
The protein FAM63A has only one known paralog: FAM63B. FAM63B is predicted as having a molecular function in the cell.[24] All of the vertebrates for which sequences are available have two copies of the FAM63 gene, both A and B. FAM63A and FAM63B likely split apart around 666 million years ago, as the closest relative to Homo sapiens containing only one FAM63 is a tapeworm, which diverged 666 million years ago.[25]
| Sequence Number | Scientific Name | Common Name | Divergence | Accession Number | Length | Identity | Similarity | Query Cover | E-value |
|---|---|---|---|---|---|---|---|---|---|
| protein FAM63B isoform a | Homo sapiens | Human | N/A | NP_001035540.1 | 621 aa | 41.9% | 76.7% | 68% | 2.00E-129 |
Expression
Promoter
The promoter region contains a number of transcription factors.[26] Those with high scores include estrogen response elements, TATA boxes, glucocoticoid response elements, and Ccaat/enchancer binding proteins. Experimental data reveals that FAM63A expression decreases when the estrogen receptor is not present, suggesting that the estrogen response elements may serve as an important promoter regulatory mechanism for this protein.[27]
Protein expression
FAM63A is a protein that is ubiquitously expressed across human tissues and throughout development. Although FAM63A is expressed ubiquitously, there are certain tissues that have higher levels of expression including the heart, thyroid, ganglia, and blood.[28]
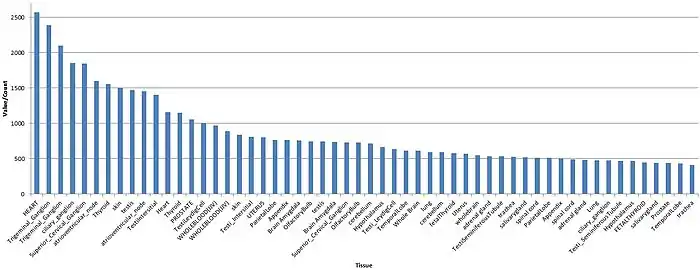
Clinical significance
Although there is no specific function determined for FAM63A, there are a few researchers who have discovered possible functions. It has been postulated that FAM63A may be associated with renal function and Chronic Kidney Disease.[8]
Figgins, Minster, and Demirci examined 17,343 functional single nucleotide polymorphisms, demonstrating a strong association between Alzheimer's Disease duration and FAM63A.[29] Another gene located on 1q21, CTSS, was also strongly associated with disease duration, the authors believe that there is a strong linkage disequilibrium between the two genes. FAM63A was identified as one of 39 genes exclusively expressed in CML cells, grouped with four other genes believed to function in protein ligation.
References
- GRCh38: Ensembl release 89: ENSG00000143409 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000038712 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Gene Cards https://www.genecards.org/cgi-bin/carddisp.pl?gene=FAM63A&search=FAM63A
- National Center for Bioinformation Technology - BLAST http://blast.ncbi.nlm.nih.gov/Blast.cgi
- Gene Cards https://www.genecards.org/cgi-bin/carddisp.pl?gene=FAM63A&search=FAM63A
- Köttgen A, Pattaro C, Böger CA, Fuchsberger C, Olden M, Glazer NL, et al. (May 2010). "New loci associated with kidney function and chronic kidney disease". Nature Genetics. 42 (5): 376–84. doi:10.1038/ng.568. PMC 2997674. PMID 20383146.
- Gene Cards https://www.genecards.org/cgi-bin/carddisp.pl?gene=FAM63A&search=FAM63A
- "Ubiquitin carboxyl-terminal hydrolase MINDY-1 isoform 1 [Homo sapiens] - Protein - NCBI".
- National Center for Bioinformation Technology - Protein https://www.ncbi.nlm.nih.gov/protein/?term=FAM63A%20AND%20homo%20sapiens
- National Center for Biotechnology Information - AceView - https://www.ncbi.nlm.nih.gov/ieb/research/acembly/av.cgi?db=human&term=FAM63A&submit=Go
- National Center for Biotechnology Information - AceView - https://www.ncbi.nlm.nih.gov/ieb/research/acembly/av.cgi?db=human&term=FAM63A&submit=Go
- National Center for Bioinformation Technology - BLAST http://blast.ncbi.nlm.nih.gov/Blast.cgi
- San Diego Super Computer http://seqtool.sdsc.edu/CGI/BW.cgi#%5B%5D!
- PSORT II Prediction http://psort.hgc.jp/form2.html
- San Diego Super Computer - http://seqtool.sdsc.edu/CGI/BW.cgi#%5B%5D!
- San Diego Super Computer - http://seqtool.sdsc.edu/CGI/BW.cgi#%5B%5D!
- ExPASy Bioinformatics Resource Portal http://www.expasy.org/proteomics
- PHYRE2 -
- Gene Cards https://www.genecards.org/cgi-bin/carddisp.pl?gene=FAM63A&search=FAM63A
- STRING - Known and Predicted Protein-Protein Interactions http://string-db.org/newstring_cgi/show_network_section.pl
- National Center for Biotechnology Information - Protein https://www.ncbi.nlm.nih.gov/protein/?term=FAM63A
- Gene Cards https://www.genecards.org/cgi-bin/carddisp.pl?gene=FAM63B
- National Center for Biotechnology Information - Protein https://www.ncbi.nlm.nih.gov/protein/?term=FAM63A
- Genomatrix - ElDorado - Promoter and transcription factors for FAM63A. http://www.genomatix.de/?s=23e6b2edc9ca33fe998f299bafe56b99
- Estrogen receptor alpha-silenced MCF7 breast cancer cells. Profile: GDS4061/ FAM63A. ncbi.nlm.nih.gov/geo/tools/profiles
- National Center for Biotechnology Information - GEO Profiles https://www.ncbi.nlm.nih.gov/geo/tools/profileGraph.cgi?ID=GDS596:221856_s_at
- Figgins JA, Minster RL, Demirci FY, Dekosky ST, Kamboh MI (June 2009). "Association studies of 22 candidate SNPs with late-onset Alzheimer's disease". American Journal of Medical Genetics. Part B, Neuropsychiatric Genetics. 150B (4): 520–6. doi:10.1002/ajmg.b.30851. PMC 2751631. PMID 18780302.