Gram domain containing 1b
GRAM domain containing 1B, also known as GRAMD1B, Aster-B and KIAA1201, is a protein that is encoded by the GRAMD1B gene.[5][6] It contains a transmembrane region and two domains of known function; the GRAM domain and a VASt domain. It is predicted to localize in the nucleus, supported by several nuclear transport signals and nuclearly associated motifs. This highly conserved gene is found in a variety of vertebrates and invertebrates, however is not found in bacteria or fungi.[7]
Gene
GRAMD1B, also known as KIAA1201, is located in the human genome at 11q24.1.[8] It is located on the + strand and is flanked by a variety of other genes. It spans 269,347 bases.[5]

mRNA
The most verified isoform, isoform 1, contains 21 exons. There are four validated isoform variants of human GRAMD1B.[5] These consist of truncated 5’ and 3’ regions, resulting in the loss of an exon.
| Isoform | mRNA length (bp) | Exons | Protein length (aa) | Status |
| 1 | 7927 | 21 | 745 | Validated |
| 2 | 7906 | 20 | 738 | Validated |
| 3 | 7636 | 20 | 698 | Validated |
| 4 | 7561 | 20 | 694 | Validated |
Protein
GRAMD1B contains several domains, motifs and signals.
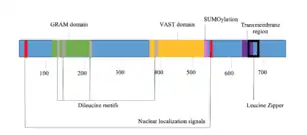
Domains
There are two confirmed cytoplasmic domains within GRAMD1B. The protein gets its name from the GRAM domain, located approximately 100 amino acids from the start codon. The GRAM domain is commonly found in myotubularin family phosphatases and predominantly involved in membrane coupled processes.[9] GRAMD1B also contains the VASt (VAD1 Analog of StAR-related lipid transfer) domain. The VASt domain is predominantly associated with lipid binding domains, such as GRAM. It is most likely to function in binding large hydrophobic ligands and may be specific for sterol.[10] A C-terminal domain in GRAMD1B sits within the lumen of the ER, is predicted to have alpha-helical secondary structure,[11] and is modified by tryptophan C-mannosyaltion.[12]
Composition Features
There are two negative charge clusters, located from amino acids 232-267 and 348-377.[13] The first cluster is not highly conserved, nor is it located in a motif or domain. The second cluster is located directly before the VASt domain and is conserved.
There are three repeat sequence regions, all fairly conserved in orthologs.[13]
| Repeat # | Sets of Repeats | Length (aa) | Location | Similarity Score |
| 1 | 3 | 18 | Within first 100 amino acids | 83.44 |
| 2 | 2 | 21 | GRAM domain | 77.22 |
| 3 | 2 | 22 | VASt domain | 67.94 |
Molecular weight and isoelectric point are conserved in orthologs.
| Region | Amino Acids[5] | Isoelectric point[14] | Molecular Weight (kdal)[15] |
| Human GRAMD1B | 745 | pH of 6.02 | 86.5 |
| GRAM domain | 94 | pH of 8.27 | 11.3 |
| VASt domain | 144 | pH of 9.41 | 17.3 |
| Transmembrane region | 21 | pH of 5.18 | 2.3 |
Structure
The mature protein contains two nuclear localization signals, pat4 and pat7.[16] There are four dileucine motifs, three located within or close to the GRAM domain.[16] A leucine zipper pattern extends through a majority the transmembrane region.[16] A SUMOylation site is located directly after the VASt domain.[17] The proteins secondary structure consists of alpha-helices, beta-strands and coils.[18] Beta-strands are mainly located within the two domains, while the alpha-helixes are concentrated near the transmembrane region. Three disulfide bonds are predicted throughout the protein.[19]
Subcellular location
GRAMD1B is predicted to localize in the nucleus and contains a transmembrane domain, most likely placing it in the membrane of the nucleus.[13]
Expression
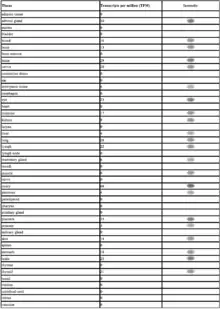
GRAMD1B is expressed in a variety of tissues. It is most highly expressed in the gonadal tissue, adrenal gland, brain and placenta.[21] It has raised expression rates in adrenal tumors, lung tumors. Developmentally, it is most highly expressed during infancy. The EST profile is supported with experimental data from multiple sources[22]

Homology
Orthologs
The ortholog space for GRAMD1B spans a large portion of evolutionary time. GRAMD1B can be found in mammals, bird, fish and invertebrates. It is not present in bacteria, archaea or fungi.[7]
| Genus species | Common Name | Accession Number | Date of Divergence (MYA)[23] | Identity[7] |
| Homo sapiens | Human | NP_001273492.1 | 0 | 100.00% |
| Aotus nancymaae | Nancy Ma's Night Monkey | XP_012325676.1 | 3.2 | 99.00% |
| Papio anubis | Olive Baboon | XP_017804515.1 | 29.44 | 97.00% |
| Castor canadensis | Beaver | XP_020037170.1 | 90 | 98.00% |
| Octodon degus | Dengu | XP_004636450.1 | 90 | 97.00% |
| Pantholops hodgsonii | Tibetan Antelope | XP_005958036.1 | 96 | 99.00% |
| Bos mutus | Domestic Yak | XP_005896826.1 | 96 | 98.00% |
| Tursiops truncatus | Dolphin | XP_019797543 | 96 | 83.00% |
| Elephantulus edwardii | Cape Elephant Shrew | XP_006895663.1 | 105 | 98.00% |
| Gallus gallus | Chicken | XP_015153638.1 | 312 | 93.00% |
| Calypte anna | Anna's Humming Bird | XP_008490701.1 | 312 | 91.00% |
| Pygoscelis adeliae | Adelie Penguin | XP_009331694.1 | 312 | 91.00% |
| Coturnix japonica | Japanese Quail | XP_015739426.1 | 312 | 90.00% |
| Anolis carolinensis | Carolina Anole | XP_008111963.1 | 312 | 87.00% |
| Danio rerio | Zebra Fish | XP_009303888.1 | 435 | 73.00% |
| Callorhinchus milii | Australian Ghost Shark | XP_007894251.1 | 473 | 77.00% |
| Branchiostoma belcheri | Lancelet | XP_019624725.1 | 684 | 40.00% |
| Octopus bimaculoides | California Two-Spot Octopus | XP_014769036.1 | 797 | 40.00% |
| Lingula anatina | Brachiopod | XP_013415578 | 797 | 38.00% |
| Zootermopsis nevadensis | Termite | KDR17240.1 | 797 | 37.00% |
| Trachymyrmex cornetzi | Ant | XP_018362289.1 | 797 | 34.00% |
Paralogs
There are four paralogs of GRAMD1B.[7] The most closely related is GRAMD1A while the most distant ortholog is GRAMD2A/GRAMD2.
| Paralog | Sequence Length | Sequence Identity[7] | Date of Divergence (MYA)[23] |
| GRAMD1A | 724 aa | 46.60% | 421.0 |
| GRAMD1C | 662 aa | 37.90% | 934.7 |
| GRAMD2B/GRAMD3 | 491 aa | 18.50% | 1625.6 |
| GRAMD2A | 353 aa | 16.70% | 1724.2 |
Phylogeny
GRAMD2 diverged earliest in history while the most recent split is GRAMD1A. The GRAMD1B gene’s rate of divergence significantly faster than Fibrinogen but is not as high as Cytochrome C.
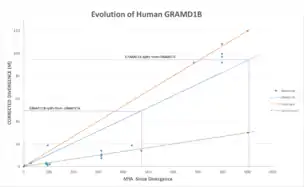
Function
Currently, the function of GRAMD1B has not been characterized.
Protein interactions
Several different proteins have been experimentally confirmed or predicted to interact with GRAMD1B.[24][25]
| Protein | Interaction identified via[24][25] | Function | Location |
| COPA | Experimental | Binds dilysine motifs. Required for budding from the Golgi and retrograde Golgi to ER transport | systolic |
| SPICE1 | Data Mining | Spindle and centriole associated. Regulates centriole duplication, proper bipolar spindle formation and chromosome congregation in mitosis | nuclear |
| GTPBP8 | Data Mining | GTP binding protein | unconfirmed |
| Ywhae | Co-sedimentation | Adapter protein associated with regulating nuclear transport to the cytoplasm | nuclear |
Clinical significance
A study tagging SNPs from chronic lymphocytic leukemia found GRAMD1B to be the second strongest risk allele region.[26] This association is supported through a number of studies[27][28] The aberrant tri-methylation of histone H3 lysine 27 induces inflammation and has been shown to increase GRAMD1B levels in colon tumors.[29]
References
- GRCh38: Ensembl release 89: ENSG00000023171 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000040111 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Entrez Gene: GRAM domain containing 1B". Retrieved 2017-02-19.
- "UniProtKB - Q3KR37 (ASTRB_HUMAN)". Retrieved March 6, 2020.
- "BLAST: Basic Local Alignment Search Tool". blast.ncbi.nlm.nih.gov. Retrieved 2 May 2017.
- "GRAMD1B Gene - GeneCards | GRM1B Protein | GRM1B Antibody". GeneCards Human Gene. Retrieved 2 May 2017.
- "GRAM Protein Domain".
- "PROSITE". prosite.expasy.org. Retrieved 2 May 2017.
- Naito T, Ercan B, Krshnan L, Triebl A, Koh DH, Wei FY, et al. (November 2019). "Movement of accessible plasma membrane cholesterol by the GRAMD1 lipid transfer protein complex". eLife. 8: e51401. doi:10.7554/eLife.51401. PMID 31724953.
- John A, Järvå MA, Shah S, Mao R, Chappaz S, Birkinshaw RW, Czabotar PE, Lo AW, Scott NE, Goddard-Borger ED (4 February 2021). "Yeast- and antibody-based tools for studying tryptophan C-mannosylation". Nature Chemical Biology. doi:10.1038/s41589-020-00727-w.
- "PSORT II Prediction". PSORT. Retrieved 2 May 2017.
- Toldo L. "Isoelectric Point Service". Archived from the original on 2008-10-26.
- "AASTATS: Statistics Based on Amino Acid Abundance, including weight and specific volume".
- "SAPS: Statistical Analysis of PS".
- "SUMOplot".
- "I-TASSER server for protein structure and function prediction". zhanglab.ccmb.med.umich.edu. Retrieved 2 May 2017.
- "DiANNA". clavius.bc.edu. Retrieved 2 May 2017.
- Remmert M. "Bioinformatics Toolkit". toolkit.tuebingen.mpg.de. Retrieved 2 May 2017.
- "EST Profile - Hs.144725". www.ncbi.nlm.nih.gov. Schuler Group. Retrieved 2 May 2017.
- "GDS1085 / 5768". www.ncbi.nlm.nih.gov. Retrieved 2 May 2017.
- "TimeTree :: The Timescale of Life". www.timetree.org. Retrieved 2 May 2017.
- "STRING: functional protein association networks". string-db.org. Retrieved 2 May 2017.
- Mike Tyers Lab. "GRAMD1B (UNQ3032/PRO9834) Result Summary | BioGRID". thebiogrid.org. Retrieved 2 May 2017.
- "OMIM Entry: 612559 - Leukemia, chronic lymphocytic, susceptibility to, 5". Online Mendelian Inheritance in Man (OMIM). Retrieved 2 May 2017.
- Lan Q, Au WY, Chanock S, Tse J, Wong KF, Shen M, Siu LP, Yuenger J, Yeager M, Hosgood HD, Purdue MP, Liang R, Rothman N (December 2010). "Genetic susceptibility for chronic lymphocytic leukemia among Chinese in Hong Kong". European Journal of Haematology. 85 (6): 492–5. doi:10.1111/j.1600-0609.2010.01518.x. PMC 2980583. PMID 20731705.
- Slager SL, Goldin LR, Strom SS, Lanasa MC, Spector LG, Rassenti L, Leis JF, Camp NJ, Kay NE, Vachon CM, Glenn M, Weinberg JB, Rabe KG, Cunningham JM, Achenbach SJ, Hanson CA, Marti GE, Call TG, Caporaso NE, Cerhan JR (April 2010). "Genetic susceptibility variants for chronic lymphocytic leukemia". Cancer Epidemiology, Biomarkers & Prevention. 19 (4): 1098–102. doi:10.1158/1055-9965.EPI-09-1217. PMC 2852480. PMID 20332261.
- Takeshima H, Ikegami D, Wakabayashi M, Niwa T, Kim YJ, Ushijima T (December 2012). "Induction of aberrant trimethylation of histone H3 lysine 27 by inflammation in mouse colonic epithelial cells". Carcinogenesis. 33 (12): 2384–90. doi:10.1093/carcin/bgs294. PMID 22976929.



