HLA-DP
HLA-DP is a protein/peptide-antigen receptor and graft-versus-host disease antigen that is composed of 2 subunits, DPα and DPβ. DPα and DPβ are encoded by two loci, HLA-DPA1 and HLA-DPB1, that are found in the MHC Class II (or HLA-D) region in the Human Leukocyte Antigen complex on human chromosome 6 (see protein boxes on right for links). Less is known about HLA-DP relative to HLA-DQ and HLA-DR but the sequencing of DP types and determination of more frequent haplotypes has progressed greatly within the last few years.
| MHC class II, DP | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| (heterodimer) | ||||||||||
| Protein type | cell surface receptor | |||||||||
| Function | Immune recognition and antigen presentation | |||||||||
| ||||||||||
Structure, Functions, Genetics
Structure
HLA-DP is an αβ-heterodimer cell-surface receptor. Each DP subunit (α-subunit, β-subunit) is composed of a α-helical N-terminal domain, an IgG-like β-sheet, a membrane spanning domain, and a cytoplasmic domain. The α-helical domain forms the sides of the peptide binding groove. The β-sheet regions form the base of the binding groove and the bulk of the molecule as well as the inter-subunit (non-covalent) binding region.
Function
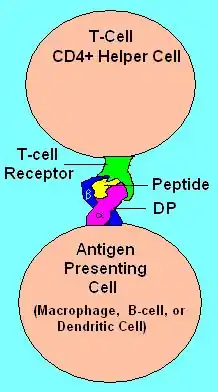
The name 'HLA-DP' originally describes a transplantation antigen of MHC class II category of the major histocompatibility complex of humans, however this antigen is an artifact of the era of organ transplantation. HLA DP functions as a cell surface receptor for foreign or self antigens. The immune system surveys antigens for foreign pathogens when presented by MHC receptors (like HLA-DP). The MHC Class II antigens are found on antigen presenting cells (APC)(macrophages, dendritic cells, and B-lymphocytes). Normally, these APC 'present' class II receptor/antigens to a great many T-cells, each with unique T-cell receptor (TCR) variants. A few TCR variants that recognize these DQ/antigen complexes are on CD4 positive T-cells. These T-cells, called T-helper (Th) cells, can promote the amplification of B-cells that recognize a different portion of the same antigen. Alternatively, macrophages and other cytotoxic lymphocytes consume or destroy cells by apoptotic signaling and present self-antigens. Self antigens, in the right context, form a regulatory T-cell population that protects self tissues from immune attack or autoimmunity.
Genetics
The α-chain and β- of DP is encoded by the HLA-DPA1 locus and HLA-DPB1 loci, respectively. This cluster is located at the proximal (centromeric) end of the HLA superlocus in human chromosome 6p21.31. It is distal from HLA-DR and HLA-DQ encoding loci and therefore is much more equilibrated with respect to other HLA loci. In the Super B8 complex DP locus is more frequently substituted, either as a result of its distance from other loci, or because it was not as actively selected in the evolution of Super B8.
Understanding the Heterodimeric DP Isoforms
| HLA | DPB1 | ||
|---|---|---|---|
| allele | (m) | (p) | |
| DPA1 | (m) | αmβm (Cis m) | αmβp (Trans) |
| (p) | αpβm (Trans) | αpβp (Cis p) | |
| Result: 2 Cis, αmβm & αpβp, isoforms and 2 trans,αmβp & αpβm. | |||
Each combination of DPA1 allele gene product with each combination of DPB1 'gene' product can potentially recombine to produce one isoform. DP genes are highly variable in the human population. In a typical population there are many DP alpha and beta. Most isoforms are not common.
These 'cis'-isoforms will account for at least 50% of the DP isoforms. The other, trans isoforms are typically more rare, isoforms result from random 'trans' combinations of haplotypes in individuals as a result of 'trans' paternal/maternal gene product isoforms.
Alleles
HLA-DPA1 Alleles
| HLA-DPA1 | |||
|---|---|---|---|
| *01 | *02 | *03 | *04 |
| *01:03 *01:04 *01:05 *01:06 *01:07 *01:08 *01:09 *01:10 |
*02:01 *02:02 *02:03 *02:04 |
*03:01 *03:02 *03:03 |
*04:01 |
HLA-DPB1 Alleles
| HLA-DPB1 | |
|---|---|
| *01 | *01:01 |
| *02 | *02:01 |
| *02:02 | |
| *03 | *03:01 |
| *04 | *04:01 |
| *04:02 | |
| *05 | *05:01 |
| *06 | *06:01 |
| *07 | *07:01 |
| *08 | *08:01 |
| *09 | *09:01 |
| *10 | *10:01 |
| DPB1*11:01 - DPB1*129:01 | |
HLA-DPB1 Allele Nomenclature Change
Before the April 2010 HLA nomenclature update, new HLA-DPB1 allele names were assigned within the existing nomenclature system. For example, the allele discovered after HLA-DPB1*9901 was assigned as DPB1*0102, the subsequent allele was named DPB1*0202, then *0302 and so on. This name assignment was decided because of the complex genetic characteristics of DPB1 alleles compared to alleles of other HLA loci. The majority of the HLA-DPB1 alleles cannot be simply grouped together by their nucleotide sequences. This name assignment has been the most confusing system within the HLA nomenclature. In the 2010 HLA nomenclature update,[1] all DPB1 alleles, except DPB1*0202 and *0402, discovered after DPB1*9901 were reassigned with new numbers. For example, DPB1*0102 becomes DPB1*100:01 and DPB1*0203 becomes DPB1*101:01.
All renamed alleles are listed in the HLA-DPB1 Nomenclature Conversion Chart below.[2]
| Previous Names | Current Names |
|---|---|
| DPB1*0102 | DPB1*100:01 |
| DPB1*0203 | DPB1*101:01 |
| DPB1*0302 | DPB1*102:01 |
| DPB1*0403 | DPB1*103:01 |
| DPB1*0502 | DPB1*104:01 |
| DPB1*0602 | DPB1*105:01 |
| DPB1*0802 | DPB1*106:01 |
| DPB1*0902 | DPB1*107:01 |
| DPB1*1002 | DPB1*108:01 |
| DPB1*1102 | DPB1*109:01 |
| DPB1*1302 | DPB1*110:01 |
| DPB1*1402 | DPB1*111:01 |
| DPB1*1502 | DPB1*112:01 |
| DPB1*1602 | DPB1*113:01 |
| DPB1*1702 | DPB1*114:01 |
| DPB1*1802 | DPB1*115:01 |
| DPB1*1902 | DPB1*116:01 |
| DPB1*2002 | DPB1*117:01 |
| DPB1*2102 | DPB1*118:01 |
| DPB1*2202 | DPB1*119:01 |
| DPB1*2302N | DPB1*120:01N |
| DPB1*2402 | DPB1*121:01 |
| DPB1*2502 | DPB1*122:01 |
| DPB1*2602 | DPB1*123:01 |
| DPB1*2702 | DPB1*124:01 |
| DPB1*2802 | DPB1*125:01 |
To aid in migration of data to the new nomenclature the WHO Nomenclature Committee for Factors of the HLA System has provided the IMGT/HLA Nomenclature Conversion Tool. This tool allows you to enter an HLA allele name and will provide you with both the current and new versions of the allele name. New alleles that have never been assigned with a name prior to the April 2010 update are:
| DPB1*126:01 DPB1*127:01 DPB1*128:01 DPB1*129:01 |
Common DP Haplotypes
| HLA-DPA1*01:03/DPB1*04:01 (DP401) |
| HLA-DPA1*01:03/DPB1*04:02 (DP402) |
External links
References
- Marsh, S. G. E., Albert, E. D., Bodmer, W. F., Bontrop, R. E., Dupont, B., Erlich, H. A., Fernández-Viña, M., Geraghty, D. E., Holdsworth, R., Hurley, C. K., Lau, M., Lee, K. W., Mach, B., Maiers, M., Mayr, W. R., Müller, C. R., Parham, P., Petersdorf, E. W., Sasazuki, T., Strominger, J. L., Svejgaard, A., Terasaki, P. I., Tiercy, J. M., Trowsdale, J. (2010). "Nomenclature for factors of the HLA system, 2010". Tissue Antigens. 75 (4): 291–455. doi:10.1111/j.1399-0039.2010.01466.x. PMC 2848993. PMID 20356336.CS1 maint: multiple names: authors list (link)
- ftp://ftp.ebi.ac.uk/pub/databases/imgt/mhc/hla/Nomenclature_2009.txt
