Hyperparameter optimization
In machine learning, hyperparameter optimization or tuning is the problem of choosing a set of optimal hyperparameters for a learning algorithm. A hyperparameter is a parameter whose value is used to control the learning process. By contrast, the values of other parameters (typically node weights) are learned.
The same kind of machine learning model can require different constraints, weights or learning rates to generalize different data patterns. These measures are called hyperparameters, and have to be tuned so that the model can optimally solve the machine learning problem. Hyperparameter optimization finds a tuple of hyperparameters that yields an optimal model which minimizes a predefined loss function on given independent data.[1] The objective function takes a tuple of hyperparameters and returns the associated loss.[1] Cross-validation is often used to estimate this generalization performance.[2]
Approaches
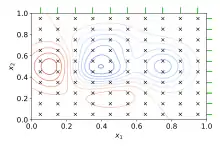
Grid search
The traditional way of performing hyperparameter optimization has been grid search, or a parameter sweep, which is simply an exhaustive searching through a manually specified subset of the hyperparameter space of a learning algorithm. A grid search algorithm must be guided by some performance metric, typically measured by cross-validation on the training set[3] or evaluation on a held-out validation set.[4]
Since the parameter space of a machine learner may include real-valued or unbounded value spaces for certain parameters, manually set bounds and discretization may be necessary before applying grid search.
For example, a typical soft-margin SVM classifier equipped with an RBF kernel has at least two hyperparameters that need to be tuned for good performance on unseen data: a regularization constant C and a kernel hyperparameter γ. Both parameters are continuous, so to perform grid search, one selects a finite set of "reasonable" values for each, say
Grid search then trains an SVM with each pair (C, γ) in the Cartesian product of these two sets and evaluates their performance on a held-out validation set (or by internal cross-validation on the training set, in which case multiple SVMs are trained per pair). Finally, the grid search algorithm outputs the settings that achieved the highest score in the validation procedure.
Grid search suffers from the curse of dimensionality, but is often embarrassingly parallel because the hyperparameter settings it evaluates are typically independent of each other.[2]
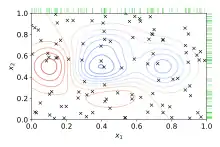
Random search
Random Search replaces the exhaustive enumeration of all combinations by selecting them randomly. This can be simply applied to the discrete setting described above, but also generalizes to continuous and mixed spaces. It can outperform Grid search, especially when only a small number of hyperparameters affects the final performance of the machine learning algorithm.[2] In this case, the optimization problem is said to have a low intrinsic dimensionality.[5] Random Search is also embarrassingly parallel, and additionally allows the inclusion of prior knowledge by specifying the distribution from which to sample.
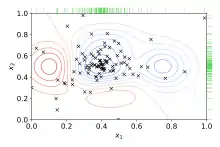
Bayesian optimization
Bayesian optimization is a global optimization method for noisy black-box functions. Applied to hyperparameter optimization, Bayesian optimization builds a probabilistic model of the function mapping from hyperparameter values to the objective evaluated on a validation set. By iteratively evaluating a promising hyperparameter configuration based on the current model, and then updating it, Bayesian optimization, aims to gather observations revealing as much information as possible about this function and, in particular, the location of the optimum. It tries to balance exploration (hyperparameters for which the outcome is most uncertain) and exploitation (hyperparameters expected close to the optimum). In practice, Bayesian optimization has been shown[6][7][8][9] to obtain better results in fewer evaluations compared to grid search and random search, due to the ability to reason about the quality of experiments before they are run.
Gradient-based optimization
For specific learning algorithms, it is possible to compute the gradient with respect to hyperparameters and then optimize the hyperparameters using gradient descent. The first usage of these techniques was focused on neural networks.[10] Since then, these methods have been extended to other models such as support vector machines[11] or logistic regression.[12]
A different approach in order to obtain a gradient with respect to hyperparameters consists in differentiating the steps of an iterative optimization algorithm using automatic differentiation.[13][14] [15]
Evolutionary optimization
Evolutionary optimization is a methodology for the global optimization of noisy black-box functions. In hyperparameter optimization, evolutionary optimization uses evolutionary algorithms to search the space of hyperparameters for a given algorithm.[7] Evolutionary hyperparameter optimization follows a process inspired by the biological concept of evolution:
- Create an initial population of random solutions (i.e., randomly generate tuples of hyperparameters, typically 100+)
- Evaluate the hyperparameters tuples and acquire their fitness function (e.g., 10-fold cross-validation accuracy of the machine learning algorithm with those hyperparameters)
- Rank the hyperparameter tuples by their relative fitness
- Replace the worst-performing hyperparameter tuples with new hyperparameter tuples generated through crossover and mutation
- Repeat steps 2-4 until satisfactory algorithm performance is reached or algorithm performance is no longer improving
Evolutionary optimization has been used in hyperparameter optimization for statistical machine learning algorithms,[7] automated machine learning, deep neural network architecture search,[16][17] as well as training of the weights in deep neural networks.[18]
Population-based
Population Based Training (PBT) learns both hyperparameter values and network weights. Multiple learning processes operate independently, using different hyperparameters. As with evolutionary methods, poorly performing models are iteratively replaced with models that adopt modified hyperparameter values and weights based on the better performers. This replacement model warm starting is the primary differentiator between PBT and other evolutionary methods. PBT thus allows the hyperparameters to evolve and eliminates the need for manual hypertuning. The process makes no assumptions regarding model architecture, loss functions or training procedures.[19]
Early stopping-based
A class of early stopping-based hyperparameter optimization algorithms is purpose built for large search spaces of continuous and discrete hyperparameters, particularly when the computational cost to evaluate the performance of a set of hyperparameters is high. Irace implements the iterated racing algorithm, that focuses the search around the most promising configurations, using statistical tests to discard the ones that perform poorly.[20][21] Another early stopping hyperparameter optimization algorithm is successive halving (SHA),[22] which begins as a random search but periodically prunes low-performing models, thereby focusing computational resources on more promising models. Asynchronous successive halving (ASHA)[23] further improves upon SHA's resource utilization profile by removing the need to synchronously evaluate and prune low-performing models. Hyperband[24] is a higher level early stopping-based algorithm that invokes SHA or ASHA multiple times with varying levels of pruning aggressiveness, in order to be more widely applicable and with fewer required inputs.
Open-source software
Grid search
- Determined, a DL Training Platform includes grid search for PyTorch and TensorFlow (Keras and Estimator) models.
- H2O AutoML provides grid search over algorithms in the H2O open source machine learning library.
- Katib is a Kubernetes-native system that includes grid search.
- scikit-learn is a Python package that includes grid search.
- Talos includes grid search for Keras.
- Tune is a Python library for distributed hyperparameter tuning and supports grid search.
Random search
- Determined is a DL Training Platform that supports random search for PyTorch and TensorFlow (Keras and Estimator) models.
- hyperopt, also via hyperas and hyperopt-sklearn, are Python packages which include random search.
- Katib is a Kubernetes-native system that includes random search.
- scikit-learn is a Python package which includes random search.
- caret is a R package which includes grid & random search.
- Talos includes a customizable random search for Keras.
- Tune is a Python library for distributed hyperparameter tuning and supports random search over arbitrary parameter distributions.
Bayesian
- Auto-sklearn[27] is a Bayesian hyperparameter optimization layer on top of scikit-learn.
- Ax[28] is a Python-based experimentation platform that supports Bayesian optimization and bandit optimization as exploration strategies.
- BOCS is a Matlab package which uses semidefinite programming for minimizing a black-box function over discrete inputs.[29] A Python 3 implementation is also included.
- HpBandSter is a Python package which combines Bayesian optimization with bandit-based methods.[30]
- Katib is a Kubernetes-native system which includes bayesian optimization.
- mlrMBO, also with mlr, is an R package for model-based/Bayesian optimization of black-box functions.
- optuna is a Python package for black box optimization, compatible with arbitrary functions that need to be optimized.
- scikit-optimize is a Python package or sequential model-based optimization with a scipy.optimize interface.[31]
- SMAC SMAC is a Python/Java library implementing Bayesian optimization.[32]
- tuneRanger is an R package for tuning random forests using model-based optimization.
Gradient-based optimization
- FAR-HO is a Python package containing Tensorflow implementations and wrappers for gradient-based hyperparamteter optimization with forward and reverse mode algorithmic differentiation.
- XGBoost is an open-source software library that provides a gradient boosting framework for C++, Java, Python, R, and Julia.
Evolutionary
- deap is a Python framework for general evolutionary computation which is flexible and integrates with parallelization packages like scoop and pyspark, and other Python frameworks like sklearn via sklearn-deap.
- Determined is a DL Training Platform that supports PBT for optimizing PyTorch and TensorFlow (Keras and Estimator) models.
- devol is a Python package that performs Deep Neural Network architecture search using genetic programming.
- nevergrad[33] is a Python package which includes Differential_evolution, Evolution_strategy, Bayesian_optimization, population control methods for the noisy case and Particle_swarm_optimization.[34]
- Tune is a Python library for distributed hyperparameter tuning and leverages nevergrad for evolutionary algorithm support.
Early Stopping
- Determined is a DL Training Platform that supports Hyperband for PyTorch and TensorFlow (Keras and Estimator) models.
- irace is an R package that implements the iterated racing algorithm.[20][21]
- Katib is a Kubernetes-native system that includes hyperband.
Other
- Determined is a DL Training Platform that supports random, grid, PBT, Hyperband and NAS approaches to hyperparameter optimization for PyTorch and TensorFlow (Keras and Estimator) models.
- dlib[35] is a C++ package with a Python API which has a parameter-free optimizer based on LIPO and trust region optimizers working in tandem.[36]
- Harmonica is a Python package for spectral hyperparameter optimization.[26]
- hyperopt, also via hyperas and hyperopt-sklearn, are Python packages which include Tree of Parzen Estimators based distributed hyperparameter optimization.
- Katib is a Kubernetes-native system which includes grid, random search, bayesian optimization, hyperband, and NAS based on reinforcement learning.
- nevergrad[33] is a Python package for gradient-free optimization using techniques such as differential evolution, sequential quadratic programming, fastGA, covariance matrix adaptation, population control methods, and particle swarm optimization.[34]
- Neural Network Intelligence (NNI) is a Python package which includes hyperparameter tuning for neural networks in local and distributed environments. Its techniques include TPE, random, anneal, evolution, SMAC, batch, grid, and hyperband.
- parameter-sherpa is a similar Python package which includes several techniques grid search, Bayesian and genetic Optimization
- photonai is a high level Python API for designing and optimizing machine learning pipelines based on grid, random search and bayesian optimization.
- pycma is a Python implementation of Covariance Matrix Adaptation Evolution Strategy.
- rbfopt is a Python package that uses a radial basis function model[25]
- Tune is a Python library for hyperparameter tuning execution and integrates with/scales many existing hyperparameter optimization libraries such as hyperopt, nevergrad, and scikit-optimize.
Commercial services
- Amazon Sagemaker uses Gaussian processes to tune hyperparameters.
- BigML OptiML supports mixed search domains
- Google HyperTune supports mixed search domains
- Indie Solver supports multiobjective, multifidelity and constraint optimization
- Mind Foundry OPTaaS supports mixed search domains, multiobjective, constraints, parallel optimization and surrogate models.
- SigOpt supports mixed search domains, multiobjective, multisolution, multifidelity, constraint (linear and black-box), and parallel optimization.
See also
References
- Claesen, Marc; Bart De Moor (2015). "Hyperparameter Search in Machine Learning". arXiv:1502.02127 [cs.LG].
- Bergstra, James; Bengio, Yoshua (2012). "Random Search for Hyper-Parameter Optimization" (PDF). Journal of Machine Learning Research. 13: 281–305.
- Chin-Wei Hsu, Chih-Chung Chang and Chih-Jen Lin (2010). A practical guide to support vector classification. Technical Report, National Taiwan University.
- Chicco D (December 2017). "Ten quick tips for machine learning in computational biology". BioData Mining. 10 (35): 35. doi:10.1186/s13040-017-0155-3. PMC 5721660. PMID 29234465.
- Ziyu, Wang; Frank, Hutter; Masrour, Zoghi; David, Matheson; Nando, de Feitas (2016). "Bayesian Optimization in a Billion Dimensions via Random Embeddings". Journal of Artificial Intelligence Research. 55: 361–387. arXiv:1301.1942. doi:10.1613/jair.4806.
- Hutter, Frank; Hoos, Holger; Leyton-Brown, Kevin (2011), "Sequential model-based optimization for general algorithm configuration" (PDF), Learning and Intelligent Optimization, Lecture Notes in Computer Science, 6683: 507–523, CiteSeerX 10.1.1.307.8813, doi:10.1007/978-3-642-25566-3_40, ISBN 978-3-642-25565-6
- Bergstra, James; Bardenet, Remi; Bengio, Yoshua; Kegl, Balazs (2011), "Algorithms for hyper-parameter optimization" (PDF), Advances in Neural Information Processing Systems
- Snoek, Jasper; Larochelle, Hugo; Adams, Ryan (2012). "Practical Bayesian Optimization of Machine Learning Algorithms" (PDF). Advances in Neural Information Processing Systems. arXiv:1206.2944. Bibcode:2012arXiv1206.2944S.
- Thornton, Chris; Hutter, Frank; Hoos, Holger; Leyton-Brown, Kevin (2013). "Auto-WEKA: Combined selection and hyperparameter optimization of classification algorithms" (PDF). Knowledge Discovery and Data Mining. arXiv:1208.3719. Bibcode:2012arXiv1208.3719T.
- Larsen, Jan; Hansen, Lars Kai; Svarer, Claus; Ohlsson, M (1996). "Design and regularization of neural networks: the optimal use of a validation set" (PDF). Proceedings of the 1996 IEEE Signal Processing Society Workshop: 62–71. CiteSeerX 10.1.1.415.3266. doi:10.1109/NNSP.1996.548336. ISBN 0-7803-3550-3.
- Olivier Chapelle; Vladimir Vapnik; Olivier Bousquet; Sayan Mukherjee (2002). "Choosing multiple parameters for support vector machines" (PDF). Machine Learning. 46: 131–159. doi:10.1023/a:1012450327387.
- Chuong B; Chuan-Sheng Foo; Andrew Y Ng (2008). "Efficient multiple hyperparameter learning for log-linear models" (PDF). Advances in Neural Information Processing Systems 20.
- Domke, Justin (2012). "Generic Methods for Optimization-Based Modeling" (PDF). Aistats. 22.
- Maclaurin, Douglas; Duvenaud, David; Adams, Ryan P. (2015). "Gradient-based Hyperparameter Optimization through Reversible Learning". arXiv:1502.03492 [stat.ML].
- Franceschi, Luca; Donini, Michele; Frasconi, Paolo; Pontil, Massimiliano (2017). "Forward and Reverse Gradient-Based Hyperparameter Optimization" (PDF). Proceedings of the 34th International Conference on Machine Learning. arXiv:1703.01785. Bibcode:2017arXiv170301785F.
- Miikkulainen R, Liang J, Meyerson E, Rawal A, Fink D, Francon O, Raju B, Shahrzad H, Navruzyan A, Duffy N, Hodjat B (2017). "Evolving Deep Neural Networks". arXiv:1703.00548 [cs.NE].
- Jaderberg M, Dalibard V, Osindero S, Czarnecki WM, Donahue J, Razavi A, Vinyals O, Green T, Dunning I, Simonyan K, Fernando C, Kavukcuoglu K (2017). "Population Based Training of Neural Networks". arXiv:1711.09846 [cs.LG].
- Such FP, Madhavan V, Conti E, Lehman J, Stanley KO, Clune J (2017). "Deep Neuroevolution: Genetic Algorithms Are a Competitive Alternative for Training Deep Neural Networks for Reinforcement Learning". arXiv:1712.06567 [cs.NE].
- Li, Ang; Spyra, Ola; Perel, Sagi; Dalibard, Valentin; Jaderberg, Max; Gu, Chenjie; Budden, David; Harley, Tim; Gupta, Pramod (2019-02-05). "A Generalized Framework for Population Based Training". arXiv:1902.01894 [cs.AI].
- López-Ibáñez, Manuel; Dubois-Lacoste, Jérémie; Pérez Cáceres, Leslie; Stützle, Thomas; Birattari, Mauro (2016). "The irace package: Iterated Racing for Automatic Algorithm Configuration". Operations Research Perspective (3): 43–58. doi:10.1016/j.orp.2016.09.002.
- Birattari, Mauro; Stützle, Thomas; Paquete, Luis; Varrentrapp, Klaus (2002). "A Racing Algorithm for Configuring Metaheuristics". GECCO 2002: 11–18.
- Jamieson, Kevin; Talwalkar, Ameet (2015-02-27). "Non-stochastic Best Arm Identification and Hyperparameter Optimization". arXiv:1502.07943 [cs.LG].
- Li, Liam; Jamieson, Kevin; Rostamizadeh, Afshin; Gonina, Ekaterina; Hardt, Moritz; Recht, Benjamin; Talwalkar, Ameet (2020-03-16). "A System for Massively Parallel Hyperparameter Tuning". arXiv:1810.05934v5.
- Li, Lisha; Jamieson, Kevin; DeSalvo, Giulia; Rostamizadeh, Afshin; Talwalkar, Ameet (2020-03-16). "Hyperband: A Novel Bandit-Based Approach to Hyperparameter Optimization". arXiv:1603.06560v4.
- Diaz, Gonzalo; Fokoue, Achille; Nannicini, Giacomo; Samulowitz, Horst (2017). "An effective algorithm for hyperparameter optimization of neural networks". arXiv:1705.08520 [cs.AI].
- Hazan, Elad; Klivans, Adam; Yuan, Yang (2017). "Hyperparameter Optimization: A Spectral Approach". arXiv:1706.00764 [cs.LG].
- Feurer M, Klein A, Eggensperger K, Springenberg J, Blum M, Hutter F (2015). "Efficient and Robust Automated Machine Learning". Advances in Neural Information Processing Systems 28 (NIPS 2015): 2962–2970.
- "Open-sourcing Ax and BoTorch: New AI tools for adaptive experimentation". 2019.
- Baptista, Ricardo; Poloczek, Matthias (2018). "Bayesian Optimization of Combinatorial Structures". arXiv:1806.08838 [stat.ML].
- Falkner, Stefan; Klein, Aaron; Hutter, Frank (2018). "BOHB: Robust and Efficient Hyperparameter Optimization at Scale". arXiv:1807.01774 [stat.ML].
- "skopt API documentation". scikit-optimize.github.io.
- Hutter F, Hoos HH, Leyton-Brown K. "Sequential Model-Based Optimization for General Algorithm Configuration" (PDF). Proceedings of the Conference on Learning and Intelligent OptimizatioN (LION 5).
- "[QUESTION] How to use to optimize NN hyperparameters · Issue #1 · facebookresearch/nevergrad". GitHub.
- "Nevergrad: An open source tool for derivative-free optimization". December 20, 2018.
- "A toolkit for making real world machine learning and data analysis applications in C++: davisking/dlib". February 25, 2019 – via GitHub.
- King, Davis. "A Global Optimization Algorithm Worth Using".
