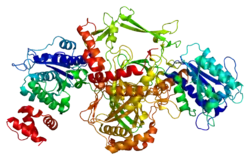
Ku80
Ku80 is a protein that, in humans, is encoded by the XRCC5 gene.[5] Together, Ku70 and Ku80 make up the Ku heterodimer, which binds to DNA double-strand break ends and is required for the non-homologous end joining (NHEJ) pathway of DNA repair. It is also required for V(D)J recombination, which utilizes the NHEJ pathway to promote antigen diversity in the mammalian immune system.
In addition to its role in NHEJ, Ku is required for telomere length maintenance and subtelomeric gene silencing.[6]
Ku was originally identified when patients with systemic lupus erythematosus were found to have high levels of autoantibodies to the protein.[7]
Nomenclature
Ku80 has been referred to by several names including:
- Lupus Ku autoantigen protein p80
- ATP-dependent DNA helicase 2 subunit 2
- X-ray repair complementing defective repair in Chinese hamster cells 5
- X-ray repair cross-complementing 5 (XRCC5)
Epigenetic repression
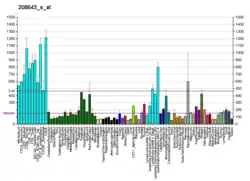
The protein expression level of Ku80 can be repressed by epigenetic hypermethylation of the promoter region of gene XRCC5 which encodes Ku80.[8] In a study of 87 matched pairs of primary tumors of non-small-cell lung carcinoma and nearby normal lung tissue, 25% of the tumors had loss of heterozygosity at the XRCC5 locus and a similar percentage of tumors had hypermethylation of the promoter region of XRCC5. Low protein expression of Ku80 was significantly associated with low mRNA expression and with XRCC5 promoter hypermethylation but not with LOH of the gene.[8]
Senescence
Mouse mutants with homozygous defects in Ku80 experience an early onset of senescence.[9][10] Ku80(-/-) mice exhibit aging-related pathology (osteopenia, atrophic skin, hepatocellular degeneration, hepatocellular inclusions, hepatic hyperplastic foci and age-specific mortality). Furthermore, Ku80(-/-) mice exhibit severely reduced lifespan and size. Loss of only a single Ku80 allele in Ku(-/+) heterozygous mice causes accelerated aging in skeletal muscle, although post natal growth is normal.[11] An analysis of the level of Ku80 protein in human, cow, and mouse indicated that Ku80 levels vary dramatically between species, and that these levels are strongly correlated with species longevity.[12] These results suggest that the NHEJ pathway of DNA repair mediated by Ku80 plays a significant role in repairing double-strand breaks that would otherwise cause early senescence (see DNA damage theory of aging).
Clinical significance
A rare microsatellite polymorphism in this gene is associated with cancer in patients of varying radiosensitivity.[5]
Deficiency in cancer
A deficiency in expression of a DNA repair gene increases the risk for cancer (see Deficient DNA repair in carcinogenesis). Ku80 protein expression was found to be deficient in melanoma.[13] In addition, low expression of Ku80 was found in 15% of adenocarcinoma type and 32% of squamous cell type non-small cell lung cancers, and this was correlated with hypermethylation of the XRCC5 promoter.[8]
Ku80 appears to be one of 26 different DNA repair proteins that are epigenetically repressed in various cancers (see Cancer epigenetics).
Interactions
Ku80 has been shown to interact with:
References
- GRCh38: Ensembl release 89: ENSG00000079246 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000026187 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Entrez Gene: XRCC5 X-ray repair complementing defective repair in Chinese hamster cells 5 (double-strand-break rejoining; Ku autoantigen, 80kDa)".
- Boulton SJ, Jackson SP (March 1998). "Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing". EMBO J. 17 (6): 1819–28. doi:10.1093/emboj/17.6.1819. PMC 1170529. PMID 9501103.
- "Entrez Gene: XRCC6 X-ray repair complementing defective repair in Chinese hamster cells 6 (Ku autoantigen, 70kDa)".
- Lee MN, Tseng RC, Hsu HS, Chen JY, Tzao C, Ho WL, Wang YC (2007). "Epigenetic inactivation of the chromosomal stability control genes BRCA1, BRCA2, and XRCC5 in non-small cell lung cancer". Clin. Cancer Res. 13 (3): 832–8. doi:10.1158/1078-0432.CCR-05-2694. PMID 17289874.
- Vogel H, Lim DS, Karsenty G, Finegold M, Hasty P (1999). "Deletion of Ku86 causes early onset of senescence in mice". Proc. Natl. Acad. Sci. U.S.A. 96 (19): 10770–5. doi:10.1073/pnas.96.19.10770. PMC 17958. PMID 10485901.
- Reiling E, Dollé ME, Youssef SA, Lee M, Nagarajah B, Roodbergen M, de With P, de Bruin A, Hoeijmakers JH, Vijg J, van Steeg H, Hasty P (2014). "The progeroid phenotype of Ku80 deficiency is dominant over DNA-PKCS deficiency". PLOS ONE. 9 (4): e93568. doi:10.1371/journal.pone.0093568. PMC 3989187. PMID 24740260.
- Didier N, Hourdé C, Amthor H, Marazzi G, Sassoon D (2012). "Loss of a single allele for Ku80 leads to progenitor dysfunction and accelerated aging in skeletal muscle". EMBO Mol Med. 4 (9): 910–23. doi:10.1002/emmm.201101075. PMC 3491824. PMID 22915554.
- Lorenzini A, Johnson FB, Oliver A, Tresini M, Smith JS, Hdeib M, Sell C, Cristofalo VJ, Stamato TD (2009). "Significant correlation of species longevity with DNA double strand break recognition but not with telomere length". Mech. Ageing Dev. 130 (11–12): 784–92. doi:10.1016/j.mad.2009.10.004. PMC 2799038. PMID 19896964.
- Korabiowska M, Tscherny M, Stachura J, Berger H, Cordon-Cardo C, Brinck U (2002). "Differential expression of DNA nonhomologous end-joining proteins Ku70 and Ku80 in melanoma progression". Mod. Pathol. 15 (4): 426–33. doi:10.1038/modpathol.3880542. PMID 11950917.
- Gell D, Jackson SP (September 1999). "Mapping of protein-protein interactions within the DNA-dependent protein kinase complex". Nucleic Acids Res. 27 (17): 3494–502. doi:10.1093/nar/27.17.3494. PMC 148593. PMID 10446239.
- Jin S, Kharbanda S, Mayer B, Kufe D, Weaver DT (October 1997). "Binding of Ku and c-Abl at the kinase homology region of DNA-dependent protein kinase catalytic subunit". J. Biol. Chem. 272 (40): 24763–6. doi:10.1074/jbc.272.40.24763. PMID 9312071.
- Matheos D, Ruiz MT, Price GB, Zannis-Hadjopoulos M (October 2002). "Ku antigen, an origin-specific binding protein that associates with replication proteins, is required for mammalian DNA replication". Biochim. Biophys. Acta. 1578 (1–3): 59–72. doi:10.1016/s0167-4781(02)00497-9. PMID 12393188.
- Barlev NA, Poltoratsky V, Owen-Hughes T, Ying C, Liu L, Workman JL, Berger SL (March 1998). "Repression of GCN5 histone acetyltransferase activity via bromodomain-mediated binding and phosphorylation by the Ku-DNA-dependent protein kinase complex". Mol. Cell. Biol. 18 (3): 1349–58. doi:10.1128/mcb.18.3.1349. PMC 108848. PMID 9488450.
- Yang CR, Yeh S, Leskov K, Odegaard E, Hsu HL, Chang C, Kinsella TJ, Chen DJ, Boothman DA (May 1999). "Isolation of Ku70-binding proteins (KUBs)". Nucleic Acids Res. 27 (10): 2165–74. doi:10.1093/nar/27.10.2165. PMC 148436. PMID 10219089.
- Singleton BK, Torres-Arzayus MI, Rottinghaus ST, Taccioli GE, Jeggo PA (May 1999). "The C terminus of Ku80 activates the DNA-dependent protein kinase catalytic subunit". Mol. Cell. Biol. 19 (5): 3267–77. doi:10.1128/mcb.19.5.3267. PMC 84121. PMID 10207052.
- Song K, Jung D, Jung Y, Lee SG, Lee I (September 2000). "Interaction of human Ku70 with TRF2". FEBS Lett. 481 (1): 81–5. doi:10.1016/s0014-5793(00)01958-x. PMID 10984620. S2CID 22753893.
- Ko L, Cardona GR, Chin WW (May 2000). "Thyroid hormone receptor-binding protein, an LXXLL motif-containing protein, functions as a general coactivator". Proc. Natl. Acad. Sci. U.S.A. 97 (11): 6212–7. doi:10.1073/pnas.97.11.6212. PMC 18584. PMID 10823961.
- Ko L, Chin WW (March 2003). "Nuclear receptor coactivator thyroid hormone receptor-binding protein (TRBP) interacts with and stimulates its associated DNA-dependent protein kinase". J. Biol. Chem. 278 (13): 11471–9. doi:10.1074/jbc.M209723200. PMID 12519782.
- Ohta S, Shiomi Y, Sugimoto K, Obuse C, Tsurimoto T (October 2002). "A proteomics approach to identify proliferating cell nuclear antigen (PCNA)-binding proteins in human cell lysates. Identification of the human CHL12/RFCs2-5 complex as a novel PCNA-binding protein". J. Biol. Chem. 277 (43): 40362–7. doi:10.1074/jbc.M206194200. PMID 12171929.
- Balajee AS, Geard CR (March 2001). "Chromatin-bound PCNA complex formation triggered by DNA damage occurs independent of the ATM gene product in human cells". Nucleic Acids Res. 29 (6): 1341–51. doi:10.1093/nar/29.6.1341. PMC 29758. PMID 11239001.
- Schild-Poulter C, Pope L, Giffin W, Kochan JC, Ngsee JK, Traykova-Andonova M, Haché RJ (May 2001). "The binding of Ku antigen to homeodomain proteins promotes their phosphorylation by DNA-dependent protein kinase". J. Biol. Chem. 276 (20): 16848–56. doi:10.1074/jbc.M100768200. PMID 11279128.
- O'Connor MS, Safari A, Liu D, Qin J, Songyang Z (July 2004). "The human Rap1 protein complex and modulation of telomere length". J. Biol. Chem. 279 (27): 28585–91. doi:10.1074/jbc.M312913200. PMID 15100233.
- Chai W, Ford LP, Lenertz L, Wright WE, Shay JW (December 2002). "Human Ku70/80 associates physically with telomerase through interaction with hTERT". J. Biol. Chem. 277 (49): 47242–7. doi:10.1074/jbc.M208542200. PMID 12377759.
- Adam L, Bandyopadhyay D, Kumar R (January 2000). "Interferon-alpha signaling promotes nucleus-to-cytoplasmic redistribution of p95Vav, and formation of a multisubunit complex involving Vav, Ku80, and Tyk2". Biochem. Biophys. Res. Commun. 267 (3): 692–6. doi:10.1006/bbrc.1999.1978. PMID 10673353.
- Karmakar P, Snowden CM, Ramsden DA, Bohr VA (August 2002). "Ku heterodimer binds to both ends of the Werner protein and functional interaction occurs at the Werner N-terminus". Nucleic Acids Res. 30 (16): 3583–91. doi:10.1093/nar/gkf482. PMC 134248. PMID 12177300.
- Li B, Comai L (September 2000). "Functional interaction between Ku and the werner syndrome protein in DNA end processing". J. Biol. Chem. 275 (37): 28349–52. doi:10.1074/jbc.C000289200. PMID 10880505.
Further reading
- Koike M (2003). "Dimerization, translocation and localization of Ku70 and Ku80 proteins". J. Radiat. Res. 43 (3): 223–36. doi:10.1269/jrr.43.223. PMID 12518983.
- Chen DJ, Park MS, Campbell E, Oshimura M, Liu P, Zhao Y, White BF, Siciliano MJ (1992). "Assignment of a human DNA double-strand break repair gene (XRCC5) to chromosome 2". Genomics. 13 (4): 1088–94. doi:10.1016/0888-7543(92)90023-L. PMID 1505945.
- Wedrychowski A, Henzel W, Huston L, Paslidis N, Ellerson D, McRae M, Seong D, Howard OM, Deisseroth A (1992). "Identification of proteins binding to interferon-inducible transcriptional enhancers in hematopoietic cells". J. Biol. Chem. 267 (7): 4533–40. PMID 1537839.
- Jeggo PA, Hafezparast M, Thompson AF, Broughton BC, Kaur GP, Zdzienicka MZ, Athwal RS (1992). "Localization of a DNA repair gene (XRCC5) involved in double-strand-break rejoining to human chromosome 2". Proc. Natl. Acad. Sci. U.S.A. 89 (14): 6423–7. doi:10.1073/pnas.89.14.6423. PMC 49513. PMID 1631138.
- Knuth MW, Gunderson SI, Thompson NE, Strasheim LA, Burgess RR (1990). "Purification and characterization of proximal sequence element-binding protein 1, a transcription activating protein related to Ku and TREF that binds the proximal sequence element of the human U1 promoter". J. Biol. Chem. 265 (29): 17911–20. PMID 2211668.
- Stuiver MH, Coenjaerts FE, van der Vliet PC (1990). "The autoantigen Ku is indistinguishable from NF IV, a protein forming multimeric protein-DNA complexes". J. Exp. Med. 172 (4): 1049–54. doi:10.1084/jem.172.4.1049. PMC 2188621. PMID 2212941.
- Mimori T, Ohosone Y, Hama N, Suwa A, Akizuki M, Homma M, Griffith AJ, Hardin JA (1990). "Isolation and characterization of cDNA encoding the 80-kDa subunit protein of the human autoantigen Ku (p70/p80) recognized by autoantibodies from patients with scleroderma-polymyositis overlap syndrome". Proc. Natl. Acad. Sci. U.S.A. 87 (5): 1777–81. doi:10.1073/pnas.87.5.1777. PMC 53566. PMID 2308937.
- Yaneva M, Wen J, Ayala A, Cook R (1989). "cDNA-derived amino acid sequence of the 86-kDa subunit of the Ku antigen". J. Biol. Chem. 264 (23): 13407–11. PMID 2760028.
- Genersch E, Eckerskorn C, Lottspeich F, Herzog C, Kühn K, Pöschl E (1995). "Purification of the sequence-specific transcription factor CTCBF, involved in the control of human collagen IV genes: subunits with homology to Ku antigen". EMBO J. 14 (4): 791–800. doi:10.1002/j.1460-2075.1995.tb07057.x. PMC 398145. PMID 7882982.
- Tuteja N, Tuteja R, Ochem A, Taneja P, Huang NW, Simoncsits A, Susic S, Rahman K, Marusic L, Chen J (1994). "Human DNA helicase II: a novel DNA unwinding enzyme identified as the Ku autoantigen". EMBO J. 13 (20): 4991–5001. doi:10.1002/j.1460-2075.1994.tb06826.x. PMC 395441. PMID 7957065.
- Le Romancer M, Reyl-Desmars F, Cherifi Y, Pigeon C, Bottari S, Meyer O, Lewin MJ (1994). "The 86-kDa subunit of autoantigen Ku is a somatostatin receptor regulating protein phosphatase-2A activity". J. Biol. Chem. 269 (26): 17464–8. PMID 8021251.
- Cao QP, Pitt S, Leszyk J, Baril EF (1994). "DNA-dependent ATPase from HeLa cells is related to human Ku autoantigen". Biochemistry. 33 (28): 8548–57. doi:10.1021/bi00194a021. PMID 8031790.
- Taccioli GE, Gottlieb TM, Blunt T, Priestley A, Demengeot J, Mizuta R, Lehmann AR, Alt FW, Jackson SP, Jeggo PA (1994). "Ku80: product of the XRCC5 gene and its role in DNA repair and V(D)J recombination". Science. 265 (5177): 1442–5. doi:10.1126/science.8073286. PMID 8073286.
- Chen DJ, Marrone BL, Nguyen T, Stackhouse M, Zhao Y, Siciliano MJ (1994). "Regional assignment of a human DNA repair gene (XRCC5) to 2q35 by X-ray hybrid mapping". Genomics. 21 (2): 423–7. doi:10.1006/geno.1994.1287. PMID 8088837.
- Maruyama K, Sugano S (1994). "Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides". Gene. 138 (1–2): 171–4. doi:10.1016/0378-1119(94)90802-8. PMID 8125298.
- Kaczmarski W, Khan SA (1993). "Lupus autoantigen Ku protein binds HIV-1 TAR RNA in vitro". Biochem. Biophys. Res. Commun. 196 (2): 935–42. doi:10.1006/bbrc.1993.2339. PMID 8240370.
- Blunt T, Taccioli GE, Priestley A, Hafezparast M, McMillan T, Liu J, Cole CC, White J, Alt FW, Jackson SP (1996). "A YAC contig encompassing the XRCC5 (Ku80) DNA repair gene and complementation of defective cells by YAC protoplast fusion". Genomics. 30 (2): 320–8. doi:10.1006/geno.1995.9871. PMID 8586433.
- Warriar N, Pagé N, Govindan MV (1996). "Expression of human glucocorticoid receptor gene and interaction of nuclear proteins with the transcriptional control element". J. Biol. Chem. 271 (31): 18662–71. doi:10.1074/jbc.271.31.18662. PMID 8702520.
- Myung K, He DM, Lee SE, Hendrickson EA (1997). "KARP-1: a novel leucine zipper protein expressed from the Ku86 autoantigen locus is implicated in the control of DNA-dependent protein kinase activity". EMBO J. 16 (11): 3172–84. doi:10.1093/emboj/16.11.3172. PMC 1169935. PMID 9214634.
- Jin S, Kharbanda S, Mayer B, Kufe D, Weaver DT (1997). "Binding of Ku and c-Abl at the kinase homology region of DNA-dependent protein kinase catalytic subunit". J. Biol. Chem. 272 (40): 24763–6. doi:10.1074/jbc.272.40.24763. PMID 9312071.
External links
- PDBe-KB provides an overview of all the structure information available in the PDB for Human X-ray repair cross-complementing protein 5