L-isoaspartyl methyltransferase
Protein L-isoaspartyl methyltransferase (PIMT, PCMT), also called S-adenosyl-L-methionine:protein-L-isoaspartate O-methyltransferase, is an enzyme which recognizes and catalyzes the repair of damaged L-isoaspartyl and D-aspartyl groups in proteins. It is a highly conserved enzyme which is present in nearly all eukaryotes, archaebacteria, and Gram-negative eubacteria.[1]
| Protein-L-isoaspartate (D-aspartate) O-methyltransferase (PIMT, PCMT) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Crystallographic structure of Human L-isoaspartyl methyltransferase.[1] | |||||||||
| Identifiers | |||||||||
| EC number | 2.1.1.77 | ||||||||
| CAS number | 105638-50-4 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
| Protein-L-isoaspartate(D-aspartate) O-methyltransferase | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||||
| Symbol | PCMT | ||||||||||
| Pfam | PF01135 | ||||||||||
| InterPro | IPR000682 | ||||||||||
| PROSITE | PDOC00985 | ||||||||||
| SCOP2 | 1dl5 / SCOPe / SUPFAM | ||||||||||
| |||||||||||
Function
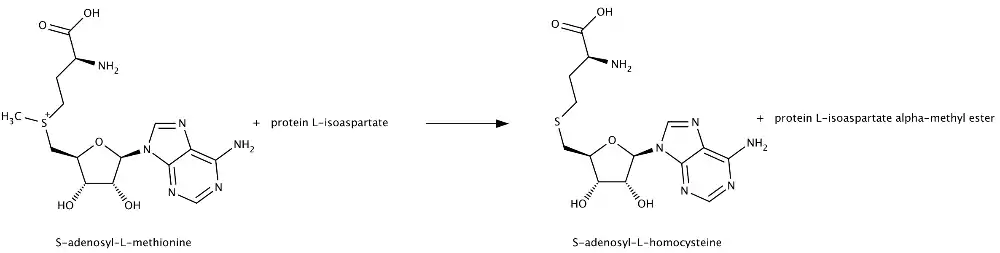
PIMT acts to transfer methyl groups from S-adenosyl-L-methionine to the alpha side chain carboxyl groups of damaged L-isoaspartyl and D-aspartyl amino acids. The enzyme takes the end methyl residue from the methionine side chain and adds it to the side chain carboxyl group of L-isoaspartate or D-aspartate to create a methyl ester. Subsequent nonenzymatic reactions result in a rapid transformation to L-succinimide, which is a precursor to aspartate and isoaspartate. The L-succinimide can then undergo nonenzymatic hydrolysis, which generates some repaired L-aspartyl residues as well as some L-isoaspartyl residues, which can then enter the cycle again for eventual conversion to the normal peptide linkage.
PIMT tends to act on proteins that have been non-enzymatically damaged due to age. By performing this repair mechanism, the enzyme helps to maintain overall protein integrity. This mechanism has been observed by several groups, and has been confirmed through experimental testing. In one report, PIMT was inhibited by adenosine dialdehyde. The results supported the proposed function of the enzyme, as the amount of abnormal L-aspartate residues increased when cells were treated with the indirect inhibitor, adenosine dialdehyde.[2] Additionally, S-adenosylhomocysteine is known to be a competitive inhibitor of PIMT.[2] When PIMT is not present in cells, the abnormal aspartyl residues accumulate, creating abnormal proteins that have been known to cause fatal progressive epilepsy in mice.[3] It has been suggested that calmodulin may play a role in stimulating the function of PIMT, although the relationship between these two molecules has not been thoroughly explored.[4] In addition to calmodulin, guanosine 5'-O-[gamma-thio]triphosphate (GTPgammaS) has been found to stimulate PIMT activity.[5]
Structure
The enzyme is present in human cytosol in two forms due to alternative splicing and differs among individuals in the population due to a single polymorphism at protein 119, either valine or isoleucine. The enzyme structure is described as a “doubly wound alpha/beta/alpha sandwich structure” which is quite consistent in all species analyzed thus far.[1] If there is any difference in the sequences between different organisms it occurs in the regions connecting the three motifs in the sandwich structure, but the sequence of the individual motifs tends to be highly conserved. Researchers have found the active site to be in the loop between the beta structure and the second alpha helix and have determined it to be highly specific for isoaspartyl residues. For example, the residues found at the C-terminus of drosophila PIMT (dPIMT) are rotated 90 degrees so as to allow more space for a substrate to interact with the enzyme. In fact, dPIMT appears to alternate between this unique open conformation and the less open conformation common of PIMT in other organisms. Although possibly unrelated to this, increased levels of dPIMT in drosophila have been correlated with increase life expectancy in these organisms due to their importance in protein repair.[6]
Reaction
See also
External links
References
- PDB: 1KR5; Ryttersgaard C, Griffith SC, Sawaya MR, MacLaren DC, Clarke S, Yeates TO (March 2002). "Crystal structure of human L-isoaspartyl methyltransferase". J. Biol. Chem. 277 (12): 10642–6. doi:10.1074/jbc.M200229200. PMID 11792715.
- Johnson BA, Najbauer J, Aswad DW (March 1993). "Accumulation of substrates for protein L-isoaspartyl methyltransferase in adenosine dialdehyde-treated PC12 cells". J. Biol. Chem. 268 (9): 6174–81. PMID 8454593.
- Yamamoto A, Takagi H, Kitamura D, Tatsuoka H, Nakano H, Kawano H, Kuroyanagi H, Yahagi Y, Kobayashi S, Koizumi K, Sakai T, Saito K, Chiba T, Kawamura K, Suzuki K, Watanabe T, Mori H, Shirasawa T (March 1998). "Deficiency in protein L-isoaspartyl methyltransferase results in a fatal progressive epilepsy". J. Neurosci. 18 (6): 2063–74. doi:10.1523/JNEUROSCI.18-06-02063.1998. PMC 6792936. PMID 9482793.
- O'Connor MB, O'Connor CM (May 1998). "Complex interactions of the protein L-isoaspartyl methyltransferase and calmodulin revealed with the yeast two-hybrid system". J. Biol. Chem. 273 (21): 12909–13. doi:10.1074/jbc.273.21.12909. PMID 9582322.
- Bilodeau D, Béliveau R (January 1999). "Inhibition of GTPgammaS-dependent L-isoaspartyl protein methylation by tyrosine kinase inhibitors in kidney". Cell. Signal. 11 (1): 45–52. doi:10.1016/S0898-6568(98)00030-8. PMID 10206344.
- Bennett EJ, Bjerregaard J, Knapp JE, Chavous DA, Friedman AM, Royer WE, O'Connor CM (November 2003). "Catalytic implications from the Drosophila protein L-isoaspartyl methyltransferase structure and site-directed mutagenesis". Biochemistry. 42 (44): 12844–53. doi:10.1021/bi034891+. PMID 14596598.