LIG3
DNA ligase 3 is an enzyme that, in humans, is encoded by the LIG3 gene.[5][6] The human LIG3 gene encodes ATP-dependent DNA ligases that seal interruptions in the phosphodiester backbone of duplex DNA.
There are three families of ATP-dependent DNA ligases in eukaryotes.[7] These enzymes utilize the same three step reaction mechanism; (i) formation of a covalent enzyme-adenylate intermediate; (ii) transfer of the adenylate group to the 5’ phosphate terminus of a DNA nick; (iii) phosphodiester bond formation. Unlike LIG1 and LIG4 family members that are found in almost all eukaryotes, LIG3 family members are less widely distributed.[8] The LIG3 gene encodes several distinct DNA ligase species by alternative translation initiation and alternative splicing mechanisms that are described below.
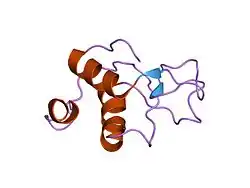
Structure, DNA binding and catalytic activities
Eukaryotic ATP-dependent DNA ligases have related catalytic region that contains three domains, a DNA binding domain, an adenylation domain and an oligonucleotide / oligosaccharide binding-fold domain. When these enzymes engage a nick in duplex DNA, these domains encircle the DNA duplex with each one making contact with the DNA. The structure of the catalytic region of DNA ligase III complexed with a nicked DNA has been determined by X-ray crystallography and is remarkably similar to that formed by the catalytic region of human DNA ligase I bound to nicked DNA.[9] A unique feature of the DNA ligases encoded by the LIG3 gene is an N-terminal zinc finger that resembles the two zinc fingers at the N-terminus of poly (ADP-ribose) polymerase 1 (PARP1).[10] As with the PARP1 zinc fingers, the DNA ligase III zinc finger is involved in binding to DNA strand breaks.[10][11][12] Within the DNA ligase III polypeptide, the zinc finger co-operates with the DNA binding domain to form a DNA binding module.[13] In addition, the adenylation domain and an oligonucleotide/oligosaccharide binding-fold domain form a second DNA binding module.[13] In the jackknife model proposed by the Ellenberger laboratory,[13] the zinc finger-DNA binding domain module serves as a strand break sensor that binds to DNA single strand interruptions irrespective of the nature of the strand break termini. If these breaks are ligatable, they are transferred to the adenylation domain-oligonucleotide/oligosaccharide binding-fold domain module that binds specifically to ligatable nicks. Compared with DNA ligases I and IV, DNA ligase III is the most active enzyme in the intermolecular joining of DNA duplexes.[14] This activity is predominantly dependent upon the DNA ligase III zinc finger suggesting that the two DNA binding modules of DNA ligase III may be able to simultaneously engage duplex DNA ends.[9][13]
Alternative splicing
The alternative translation initiation and splicing mechanisms alter the amino- and carboxy-terminal sequences that flank the DNA ligase III catalytic region.[15][16] In the alternative splicing mechanism, the exon encoding a C-terminal breast cancer susceptibility protein 1 C-terminal (BRCT) domain at the C-terminus of DNA ligase III-alpha is replaced by a short positively charged sequence that acts as a nuclear localization signal, generating DNA ligase III-beta. This alternatively spliced variant has, to date, only been detected in male germs cells.[16] Based on its expression pattern during spermatogenesis, it appears likely that DNA ligase IIIbeta is involved in meiotic recombination and/or DNA repair in haploid sperm but this has not been definitively demonstrated. Although an internal ATG is the preferred site for translation initiation within the DNA ligase III open reading frame, translation initiations does also occur at the first ATG within the open reading frame, resulting in the synthesis of a polypeptide with an N-terminal mitochondrial targeting sequence.[15][17][18]
Cellular function
As mentioned above, DNA ligase III-alpha mRNA encodes nuclear and mitochondrial versions of DNA ligase III-alpha. Nuclear DNA ligase III-alpha exists and functions in a stable complex with the DNA repair protein XRCC1.[19][20] These proteins interact via their C-terminal BRCT domains.[16][21] XRCC1 has no enzymatic activity but instead appears to acts as a scaffold protein by interacting with a large number of proteins involved in base excision and single-strand break repair. The participation of XRCC1 in these pathways is consistent with the phenotype of xrcc1 cells.[19] In contrast to nuclear DNA ligase III-alpha, mitochondrial DNA ligase III-alpha functions independently of XRCC1, which is not found in mitochondria.[22] It appears that nuclear DNA ligase III-alpha forms a complex with XRCC1 in the cytoplasm and the subsequent nuclear targeting of the resultant complex is directed by the XRCC1 nuclear localization signal.[23] While mitochondrial DNA ligase III-alpha also interacts with XRCC1, it is likely that the activity of the mitochondrial targeting sequence of DNA ligase III-alpha is greater than the activity of the XRCC1 nuclear localization signal and that the DNA ligase III-alpha/XRCC1 complex is disrupted when mitochondrial DNA ligase III-alpha passes through the mitochondrial membrane.
Since the LIG3 gene encodes the only DNA ligase in mitochondria, inactivation of the LIG3 gene results in loss of mitochondrial DNA that in turn leads to loss of mitochondrial function.[24][25][26] Fibroblasts with inactivated Lig3 gene can be propagated in the media supplemented with uridine and pyruvate. However, these cells lack mtDNA.[27] Physiological levels of mitochondrial DNA ligase III appear excessive, and cells with 100-fold reduced mitochondrial content of mitochondrial DNA ligase III-alpha maintain normal mtDNA copy number.[27] The essential role of DNA ligase III-alpha in mitochondrial DNA metabolism can be fulfilled by other DNA ligases, including the NAD-dependent DNA ligase of E. coli, if they are targeted to mitochondria.[24][26] Thus, viable cells that lack nuclear DNA ligase III-alpha can be generated. While DNA ligase I is the predominant enzyme that joins Okazaki fragments during DNA replication, it is now evident that the DNA ligase III-alpha/XRCC1 complex enables cells that either lack or have reduced DNA ligase I activity to complete DNA replication.[24][26][28][29] Given the biochemical and cell biology studies linking the DNA ligase III-alpha/XRCC1 complex with excision repair and the repair of DNA single strand breaks,[30][31][32][33] it was surprising that the cells lacking nuclear DNA ligase III-alpha did not exhibit significantly increased sensitivity to DNA damaging agent.[24][26] These studies suggest that there is significant functional redundancy between DNA ligase I and DNA ligase III-alpha in these nuclear DNA repair pathways. In mammalian cells, most DNA double strand breaks are repaired by DNA ligase IV-dependent non-homologous end joining (NHEJ).[34] DNA ligase III-alpha participates in a minor alternative NHEJ pathway that generates chromosomal translocations.[35][36] Unlike the other nuclear DNA repair functions, it appears that the role of DNA ligase III-alpha in alternative NHEJ is independent of XRCC1.[37]
Clinical significance
Unlike the LIG1 and LIG4 genes,[38][39][40][41] inherited mutations in the LIG3 gene have not been identified in the human population. DNA ligase III-alpha has, however, been indirectly implicated in cancer and neurodegenerative diseases. In cancer, DNA ligase III-alpha is frequently overexpressed and this serves as a biomarker to identify cells that are more dependent upon the alternative NHEJ pathway for the repair of DNA double strand breaks.[42][43][44][45] Although the increased activity of the alternative NHEJ pathway causes genomic instability that drives disease progression, it also constitutes a novel target for the development of cancer cell-specific therapeutic strategies.[43][44] Several genes encoding proteins that interact directly with DNA ligase III-alpha or indirectly via interactions with XRCC1 have been identified as being mutated in inherited neurodegenerative diseases.[46][47][48][49][50] Thus, it appears that DNA transactions involving DNA ligase III-alpha play an important role in maintaining the viability of neuronal cells.
LIG3 has a role in microhomology-mediated end joining (MMEJ) repair of double strand breaks. It is one of 6 enzymes required for this error prone DNA repair pathway.[51] LIG3 is upregulated in chronic myeloid leukemia,[45] multiple myeloma,[52] and breast cancer.[43]
Cancers are very often deficient in expression of one or more DNA repair genes, but over-expression of a DNA repair gene is less usual in cancer. For instance, at least 36 DNA repair enzymes, when mutationally defective in germ line cells, cause increased risk of cancer (hereditary cancer syndromes). (Also see DNA repair-deficiency disorder.) Similarly, at least 12 DNA repair genes have frequently been found to be epigenetically repressed in one or more cancers. (See also Epigenetically reduced DNA repair and cancer.) Ordinarily, deficient expression of a DNA repair enzyme results in increased un-repaired DNA damages which, through replication errors (translesion synthesis), lead to mutations and cancer. However, LIG3 mediated MMEJ repair is highly inaccurate, so in this case, over-expression, rather than under-expression, apparently leads to cancer.
Notes
References
- GRCh38: Ensembl release 89: ENSG00000005156 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000020697 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Entrez Gene: Ligase III, DNA, ATP-dependent". Retrieved 2012-03-12.
- Tomkinson AE, Sallmyr A (December 2013). "Structure and function of the DNA ligases encoded by the mammalian LIG3 gene". Gene. 531 (2): 150–7. doi:10.1016/j.gene.2013.08.061. PMC 3881560. PMID 24013086.
- Ellenberger T, Tomkinson AE (2008). "Eukaryotic DNA ligases: structural and functional insights". Annu. Rev. Biochem. 77: 313–38. doi:10.1146/annurev.biochem.77.061306.123941. PMC 2933818. PMID 18518823.
- Simsek D, Jasin M (November 2011). "DNA ligase III: a spotty presence in eukaryotes, but an essential function where tested". Cell Cycle. 10 (21): 3636–44. doi:10.4161/cc.10.21.18094. PMC 3266004. PMID 22041657.
- Cotner-Gohara E, Kim IK, Hammel M, Tainer JA, Tomkinson AE, Ellenberger T (July 2010). "Human DNA ligase III recognizes DNA ends by dynamic switching between two DNA-bound states". Biochemistry. 49 (29): 6165–76. doi:10.1021/bi100503w. PMC 2922849. PMID 20518483.
- Mackey ZB, Niedergang C, Murcia JM, Leppard J, Au K, Chen J, de Murcia G, Tomkinson AE (July 1999). "DNA ligase III is recruited to DNA strand breaks by a zinc finger motif homologous to that of poly(ADP-ribose) polymerase. Identification of two functionally distinct DNA binding regions within DNA ligase III". Journal of Biological Chemistry. 274 (31): 21679–87. doi:10.1074/jbc.274.31.21679. PMID 10419478.
- Leppard JB, Dong Z, Mackey ZB, Tomkinson AE (August 2003). "Physical and functional interaction between DNA ligase IIIalpha and poly(ADP-Ribose) polymerase 1 in DNA single-strand break repair". Molecular and Cellular Biology. 23 (16): 5919–27. doi:10.1128/MCB.23.16.5919-5927.2003. PMC 166336. PMID 12897160.
- Taylor RM, Whitehouse CJ, Caldecott KW (September 2000). "The DNA ligase III zinc finger stimulates binding to DNA secondary structure and promotes end joining". Nucleic Acids Res. 28 (18): 3558–63. doi:10.1093/nar/28.18.3558. PMC 110727. PMID 10982876.
- Cotner-Gohara E, Kim IK, Tomkinson AE, Ellenberger T (April 2008). "Two DNA-binding and nick recognition modules in human DNA ligase III". Journal of Biological Chemistry. 283 (16): 10764–72. doi:10.1074/jbc.M708175200. PMC 2447648. PMID 18238776.
- Chen L, Trujillo K, Sung P, Tomkinson AE (August 2000). "Interactions of the DNA ligase IV-XRCC4 complex with DNA ends and the DNA-dependent protein kinase". Journal of Biological Chemistry. 275 (34): 26196–205. doi:10.1074/jbc.M000491200. PMID 10854421.
- Lakshmipathy U, Campbell C (May 1999). "The human DNA ligase III gene encodes nuclear and mitochondrial proteins". Molecular and Cellular Biology. 19 (5): 3869–76. doi:10.1128/MCB.19.5.3869. PMC 84244. PMID 10207110.
- Mackey ZB, Ramos W, Levin DS, Walter CA, McCarrey JR, Tomkinson AE (February 1997). "An alternative splicing event which occurs in mouse pachytene spermatocytes generates a form of DNA ligase III with distinct biochemical properties that may function in meiotic recombination". Molecular and Cellular Biology. 17 (2): 989–98. doi:10.1128/MCB.17.2.989. PMC 231824. PMID 9001252.
- Wei YF, Robins P, Carter K, Caldecott K, Pappin DJ, Yu GL, Wang RP, Shell BK, Nash RA, Schär P (June 1995). "Molecular cloning and expression of human cDNAs encoding a novel DNA ligase IV and DNA ligase III, an enzyme active in DNA repair and recombination". Molecular and Cellular Biology. 15 (6): 3206–16. doi:10.1128/mcb.15.6.3206. PMC 230553. PMID 7760816.
- Chen J, Tomkinson AE, Ramos W, Mackey ZB, Danehower S, Walter CA, Schultz RA, Besterman JM, Husain I (October 1995). "Mammalian DNA ligase III: molecular cloning, chromosomal localization, and expression in spermatocytes undergoing meiotic recombination". Molecular and Cellular Biology. 15 (10): 5412–22. doi:10.1128/MCB.15.10.5412. PMC 230791. PMID 7565692.
- Caldecott KW, McKeown CK, Tucker JD, Ljungquist S, Thompson LH (January 1994). "An interaction between the mammalian DNA repair protein XRCC1 and DNA ligase III". Molecular and Cellular Biology. 14 (1): 68–76. doi:10.1128/MCB.14.1.68. PMC 358357. PMID 8264637.
- Caldecott KW, Tucker JD, Stanker LH, Thompson LH (December 1995). "Characterization of the XRCC1-DNA ligase III complex in vitro and its absence from mutant hamster cells". Nucleic Acids Res. 23 (23): 4836–43. doi:10.1093/nar/23.23.4836. PMC 307472. PMID 8532526.
- Nash RA, Caldecott KW, Barnes DE, Lindahl T (April 1997). "XRCC1 protein interacts with one of two distinct forms of DNA ligase III". Biochemistry. 36 (17): 5207–11. doi:10.1021/bi962281m. PMID 9136882.
- Lakshmipathy U, Campbell C (October 2000). "Mitochondrial DNA ligase III function is independent of Xrcc1". Nucleic Acids Res. 28 (20): 3880–6. doi:10.1093/nar/28.20.3880. PMC 110795. PMID 11024166.
- Parsons JL, Dianova II, Finch D, Tait PS, Ström CE, Helleday T, Dianov GL (July 2010). "XRCC1 phosphorylation by CK2 is required for its stability and efficient DNA repair". DNA Repair (Amst.). 9 (7): 835–41. doi:10.1016/j.dnarep.2010.04.008. PMID 20471329.
- Gao Y, Katyal S, Lee Y, Zhao J, Rehg JE, Russell HR, McKinnon PJ (March 2011). "DNA ligase III is critical for mtDNA integrity but not Xrcc1-mediated nuclear DNA repair". Nature. 471 (7337): 240–4. doi:10.1038/nature09773. PMC 3079429. PMID 21390131.
- Lakshmipathy U, Campbell C (February 2001). "Antisense-mediated decrease in DNA ligase III expression results in reduced mitochondrial DNA integrity". Nucleic Acids Res. 29 (3): 668–76. doi:10.1093/nar/29.3.668. PMC 30390. PMID 11160888.
- Simsek D, Furda A, Gao Y, Artus J, Brunet E, Hadjantonakis AK, Van Houten B, Shuman S, McKinnon PJ, Jasin M (March 2011). "Crucial role for DNA ligase III in mitochondria but not in Xrcc1-dependent repair". Nature. 471 (7337): 245–8. doi:10.1038/nature09794. PMC 3261757. PMID 21390132.
- Shokolenko, IN; Fayzulin, RZ; Katyal, S; McKinnon, PJ; Alexeyev, MF (Sep 13, 2013). "Mitochondrial DNA ligase is dispensable for the viability of cultured cells but essential for mtDNA maintenance". Journal of Biological Chemistry. 288 (37): 26594–605. doi:10.1074/jbc.M113.472977. PMC 3772206. PMID 23884459.
- Arakawa H, Bednar T, Wang M, Paul K, Mladenov E, Bencsik-Theilen AA, Iliakis G (March 2012). "Functional redundancy between DNA ligases I and III in DNA replication in vertebrate cells". Nucleic Acids Research. 40 (6): 2599–610. doi:10.1093/nar/gkr1024. PMC 3315315. PMID 22127868.
- Le Chalony C, Hoffschir F, Gauthier LR, Gross J, Biard DS, Boussin FD, Pennaneach V (September 2012). "Partial complementation of a DNA ligase I deficiency by DNA ligase III and its impact on cell survival and telomere stability in mammalian cells". Cellular and Molecular Life Sciences. 69 (17): 2933–49. doi:10.1007/s00018-012-0975-8. PMC 3417097. PMID 22460582.
- Cappelli E, Taylor R, Cevasco M, Abbondandolo A, Caldecott K, Frosina G (September 1997). "Involvement of XRCC1 and DNA ligase III gene products in DNA base excision repair". Journal of Biological Chemistry. 272 (38): 23970–5. doi:10.1074/jbc.272.38.23970. PMID 9295348.
- Okano S, Lan L, Tomkinson AE, Yasui A (2005). "Translocation of XRCC1 and DNA ligase III-alpha from centrosomes to chromosomes in response to DNA damage in mitotic human cells". Nucleic Acids Res. 33 (1): 422–9. doi:10.1093/nar/gki190. PMC 546168. PMID 15653642.
- Kubota Y, Nash RA, Klungland A, Schär P, Barnes DE, Lindahl T (December 1996). "Reconstitution of DNA base excision-repair with purified human proteins: interaction between DNA polymerase beta and the XRCC1 protein". EMBO Journal. 15 (23): 6662–70. doi:10.1002/j.1460-2075.1996.tb01056.x. PMC 452490. PMID 8978692.
- Moser J, Kool H, Giakzidis I, Caldecott K, Mullenders LH, Fousteri MI (July 2007). "Sealing of chromosomal DNA nicks during nucleotide excision repair requires XRCC1 and DNA ligase III alpha in a cell-cycle-specific manner" (PDF). Molecular Cell. 27 (2): 311–23. doi:10.1016/j.molcel.2007.06.014. PMID 17643379.
- Lieber MR (2010). "The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway". Annu. Rev. Biochem. 79: 181–211. doi:10.1146/annurev.biochem.052308.093131. PMC 3079308. PMID 20192759.
- Wang H, Rosidi B, Perrault R, Wang M, Zhang L, Windhofer F, Iliakis G (May 2005). "DNA ligase III as a candidate component of backup pathways of nonhomologous end joining". Cancer Res. 65 (10): 4020–30. doi:10.1158/0008-5472.CAN-04-3055. PMID 15899791.
- Simsek D, Brunet E, Wong SY, Katyal S, Gao Y, McKinnon PJ, Lou J, Zhang L, Li J, Rebar EJ, Gregory PD, Holmes MC, Jasin M (June 2011). Haber JE (ed.). "DNA ligase III promotes alternative nonhomologous end-joining during chromosomal translocation formation". PLOS Genet. 7 (6): e1002080. doi:10.1371/journal.pgen.1002080. PMC 3107202. PMID 21655080.
- Boboila C, Oksenych V, Gostissa M, Wang JH, Zha S, Zhang Y, Chai H, Lee CS, Jankovic M, Saez LM, Nussenzweig MC, McKinnon PJ, Alt FW, Schwer B (February 2012). "Robust chromosomal DNA repair via alternative end-joining in the absence of X-ray repair cross-complementing protein 1 (XRCC1)". Proceedings of the National Academy of Sciences, USA. 109 (7): 2473–8. doi:10.1073/pnas.1121470109. PMC 3289296. PMID 22308491.
- Girard PM, Kysela B, Härer CJ, Doherty AJ, Jeggo PA (October 2004). "Analysis of DNA ligase IV mutations found in LIG4 syndrome patients: the impact of two linked polymorphisms". Human Molecular Genetics. 13 (20): 2369–76. doi:10.1093/hmg/ddh274. PMID 15333585.
- O'Driscoll M, Cerosaletti KM, Girard PM, Dai Y, Stumm M, Kysela B, Hirsch B, Gennery A, Palmer SE, Seidel J, Gatti RA, Varon R, Oettinger MA, Neitzel H, Jeggo PA, Concannon P (December 2001). "DNA ligase IV mutations identified in patients exhibiting developmental delay and immunodeficiency". Molecular Cell. 8 (6): 1175–85. doi:10.1016/S1097-2765(01)00408-7. PMID 11779494.
- Riballo E, Critchlow SE, Teo SH, Doherty AJ, Priestley A, Broughton B, Kysela B, Beamish H, Plowman N, Arlett CF, Lehmann AR, Jackson SP, Jeggo PA (July 1999). "Identification of a defect in DNA ligase IV in a radiosensitive leukaemia patient". Curr. Biol. 9 (13): 699–702. doi:10.1016/S0960-9822(99)80311-X. PMID 10395545. S2CID 17103936.
- Barnes DE, Tomkinson AE, Lehmann AR, Webster AD, Lindahl T (May 1992). "Mutations in the DNA ligase I gene of an individual with immunodeficiencies and cellular hypersensitivity to DNA-damaging agents". Cell. 69 (3): 495–503. doi:10.1016/0092-8674(92)90450-Q. PMID 1581963. S2CID 11736507.
- Chen X, Zhong S, Zhu X, Dziegielewska B, Ellenberger T, Wilson GM, MacKerell AD, Tomkinson AE (May 2008). "Rational design of human DNA ligase inhibitors that target cellular DNA replication and repair". Cancer Res. 68 (9): 3169–77. doi:10.1158/0008-5472.CAN-07-6636. PMC 2734474. PMID 18451142.
- Tobin LA, Robert C, Nagaria P, Chumsri S, Twaddell W, Ioffe OB, Greco GE, Brodie AH, Tomkinson AE, Rassool FV (2012). "Targeting abnormal DNA repair in therapy-resistant breast cancers". Molecular Cancer Research. 10 (1): 96–107. doi:10.1158/1541-7786.MCR-11-0255. PMC 3319138. PMID 22112941.
- Tobin LA, Robert C, Rapoport AP, Gojo I, Baer MR, Tomkinson AE, Rassool FV (April 2013). "Targeting abnormal DNA double-strand break repair in tyrosine kinase inhibitor-resistant chronic myeloid leukemias". Oncogene. 32 (14): 1784–93. doi:10.1038/onc.2012.203. PMC 3752989. PMID 22641215.
- Sallmyr A, Tomkinson AE, Rassool FV (August 2008). "Up-regulation of WRN and DNA ligase III-alpha in chronic myeloid leukemia: consequences for the repair of DNA double-strand breaks". Blood. 112 (4): 1413–23. doi:10.1182/blood-2007-07-104257. PMC 2967309. PMID 18524993.
- Ahel I, Rass U, El-Khamisy SF, Katyal S, Clements PM, McKinnon PJ, Caldecott KW, West SC (October 2006). "The neurodegenerative disease protein aprataxin resolves abortive DNA ligation intermediates". Nature. 443 (7112): 713–6. doi:10.1038/nature05164. PMID 16964241. S2CID 4431045.
- Date H, Onodera O, Tanaka H, Iwabuchi K, Uekawa K, Igarashi S, Koike R, Hiroi T, Yuasa T, Awaya Y, Sakai T, Takahashi T, Nagatomo H, Sekijima Y, Kawachi I, Takiyama Y, Nishizawa M, Fukuhara N, Saito K, Sugano S, Tsuji S (October 2001). "Early-onset ataxia with ocular motor apraxia and hypoalbuminemia is caused by mutations in a new HIT superfamily gene". Nature Genetics. 29 (2): 184–8. doi:10.1038/ng1001-184. PMID 11586299. S2CID 25665707.
- Moreira MC, Barbot C, Tachi N, Kozuka N, Uchida E, Gibson T, Mendonça P, Costa M, Barros J, Yanagisawa T, Watanabe M, Ikeda Y, Aoki M, Nagata T, Coutinho P, Sequeiros J, Koenig M (October 2001). "The gene mutated in ataxia-ocular apraxia 1 encodes the new HIT/Zn-finger protein aprataxin". Nature Genetics. 29 (2): 189–93. doi:10.1038/ng1001-189. PMID 11586300. S2CID 23001321.
- El-Khamisy SF, Saifi GM, Weinfeld M, Johansson F, Helleday T, Lupski JR, Caldecott KW (March 2005). "Defective DNA single-strand break repair in spinocerebellar ataxia with axonal neuropathy-1". Nature. 434 (7029): 108–13. doi:10.1038/nature03314. PMID 15744309. S2CID 4423748.
- Shen J, Gilmore EC, Marshall CA, Haddadin M, Reynolds JJ, Eyaid W, Bodell A, Barry B, Gleason D, Allen K, Ganesh VS, Chang BS, Grix A, Hill RS, Topcu M, Caldecott KW, Barkovich AJ, Walsh CA (March 2010). "Mutations in PNKP cause microcephaly, seizures and defects in DNA repair". Nature Genetics. 42 (3): 245–9. doi:10.1038/ng.526. PMC 2835984. PMID 20118933.
- Sharma S, Javadekar SM, Pandey M, Srivastava M, Kumari R, Raghavan SC (2015). "Homology and enzymatic requirements of microhomology-dependent alternative end joining". Cell Death Dis. 6 (3): e1697. doi:10.1038/cddis.2015.58. PMC 4385936. PMID 25789972.
- Herrero AB, San Miguel J, Gutierrez NC (2015). "Deregulation of DNA double-strand break repair in multiple myeloma: implications for genome stability". PLOS ONE. 10 (3): e0121581. doi:10.1371/journal.pone.0121581. PMC 4366222. PMID 25790254.