Nearly neutral theory of molecular evolution
The nearly neutral theory of molecular evolution is a modification of the neutral theory of molecular evolution that accounts for the fact that not all mutations are either so deleterious such that they can be ignored, or else neutral. Slightly deleterious mutations are reliably purged only when their selection coefficient are greater than one divided by the effective population size. In larger populations, a higher proportion of mutations exceed this threshold for which genetic drift cannot overpower selection, leading to fewer fixation events and so slower molecular evolution.
The nearly neutral theory was proposed by Tomoko Ohta in 1973.[1] The population-size-dependent threshold for purging mutations has been called the "drift barrier" by Michael Lynch, and used to explain differences in genomic architecture among species.
Origins of the nearly neutral theory
According to the neutral theory of molecular evolution, the rate at which molecular changes accumulate between species should be equal to the rate of neutral mutations and hence relatively constant across species. However, this is a per-generation rate. Since larger organisms have longer generation times, the neutral theory predicts that their rate of molecular evolution should be slower. However, molecular evolutionists found that rates of protein evolution were fairly independent of generation time.
Noting that population size is generally inversely proportional to generation time, Tomoko Ohta proposed that if most amino acid substitutions are slightly deleterious, this would increase the rate of effectively neutral mutation rate in small populations, which could offset the effect of long generation times. However, because noncoding DNA substitutions tend to be more neutral, independent of population size, their rate of evolution is correctly predicted to depend on population size / generation time, unlike the rate of non-synonymous changes.[2]
In this case, the faster rate of neutral evolution in proteins expected in small populations (due to a more lenient threshold for purging deleterious mutations) is offset by longer generation times (and vice versa), but in large populations with short generation times, noncoding DNA evolves faster while protein evolution is retarded by selection (which is more significant than drift for large populations)[2] In 1973, Ohta published a short letter in Nature[1] suggesting that a wide variety of molecular evidence supported the theory that most mutation events at the molecular level are slightly deleterious rather than strictly neutral.
Between then and the early 1990s, many studies of molecular evolution used a "shift model" in which the negative effect on the fitness of a population due to deleterious mutations shifts back to an original value when a mutation reaches fixation. In the early 1990s, Ohta developed a "fixed model" that included both beneficial and deleterious mutations, so that no artificial "shift" of overall population fitness was necessary.[2] According to Ohta, however, the nearly neutral theory largely fell out of favor in the late 1980s, because the mathematically simpler neutral theory for the widespread molecular systematics research that flourished after the advent of rapid DNA sequencing. As more detailed systematics studies started to compare the evolution of genome regions subject to strong selection versus weaker selection in the 1990s, the nearly neutral theory and the interaction between selection and drift have once again become an important focus of research.[3]
Theory
The rate of substitution, is
- ,
where is the mutation rate, is the generation time, and is the effective population size. The last term is the probability that a new mutation will become fixed. Early models assumed that is constant between species, and that increases with . Kimura’s equation for the probability of fixation in a haploid population gives:
- ,
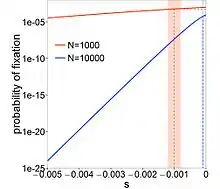
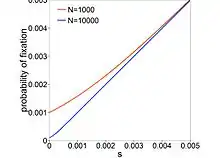
where is the selection coefficient of a mutation. When (completely neutral), , and when (extremely deleterious), decreases almost exponentially with . Mutations with are called nearly neutral mutations. These mutations can fix in small- populations through genetic drift. In large- populations, these mutations are purged by selection. If nearly neutral mutations are common, then the proportion for which is dependent on
The effect of nearly neutral mutations can depend on fluctuations in . Early work used a “shift model” in which can vary between generations but the mean fitness of the population is reset to zero after fixation. This basically assumes the distribution of is constant (in this sense, the argument in the previous paragraphs can be regarded as based on the “shift model”). This assumption can lead to indefinite improvement or deterioration of protein function. Alternatively, the later “fixed model”[4] fixes the distribution of mutations’ effect on protein function, but allows the mean fitness of population to evolve. This allows the distribution of to change with the mean fitness of population.
The “fixed model” provides a slightly different explanation for the rate of protein evolution. In large populations, advantageous mutations are quickly picked up by selection, increasing the mean fitness of the population. In response, the mutation rate of nearly neutral mutations is reduced because these mutations are restricted to the tail of the distribution of selection coefficients.
The “fixed model” expands the nearly neutral theory. Tachida[5] classified evolution under the “fixed model” based on the product of and the variance in the distribution of : a large product corresponds to adaptive evolution, an intermediate product corresponds to nearly neutral evolution, and a small product corresponds to almost neutral evolution. According to this classification, slightly advantageous mutations can contribute to nearly neutral evolution.
The "drift barrier" theory
Michael Lynch has proposed that variation in the ability to purge slightly deleterious mutations (i.e. variation in ) can explain variation in genomic architecture among species, e.g. the size of the genome, or the mutation rate.[6] Specifically, larger populations will have lower mutation rates, more streamlined genomic architectures, and generally more finely tuned adaptations. However, if robustness to the consequences of each possible error in processes such as transcription and translation substantially reduces the cost of making such errors, larger populations might evolve lower rates of global proofreading, and hence have higher rates of error.[7] This may explain why Escherichia coli has higher rates of transcription error than Saccharomyces cerevisiae.[8][9] This is supported by the fact that transcriptional error rates in E. coli depend on protein abundance (which is responsible for modulating the locus-specific strength of selection), but do so only for high-error-rate C to U deamination errors in S. cerevisiae.[10]
References
- Ohta T (November 1973). "Slightly deleterious mutant substitutions in evolution". Nature. 246 (5428): 96–8. doi:10.1038/246096a0. PMID 4585855.
- Ohta T, Gillespie JH (April 1996). "Development of Neutral and Nearly Neutral Theories". Theoretical Population Biology. 49 (2): 128–42. CiteSeerX 10.1.1.332.2080. doi:10.1006/tpbi.1996.0007. PMID 8813019.
- Ohta T (August 1996). "The current significance and standing of neutral and neutral theories". BioEssays. 18 (8): 673–7, discussion 683. doi:10.1002/bies.950180811. PMID 8779656.
- Ohta T, Tachida H (September 1990). "Theoretical study of near neutrality. I. Heterozygosity and rate of mutant substitution". Genetics. 126 (1): 219–29. PMC 1204126. PMID 2227381.
- Tachida H (May 1991). "A study on a nearly neutral mutation model in finite populations". Genetics. 128 (1): 183–92. PMC 1204447. PMID 2060776.
- Lynch M (2007). The origins of genome architecture. Sunderland: Sinauer Associates.
- Rajon, E.; Masel, J. (3 January 2011). "Evolution of molecular error rates and the consequences for evolvability". Proceedings of the National Academy of Sciences. 108 (3): 1082–1087. Bibcode:2011PNAS..108.1082R. doi:10.1073/pnas.1012918108. PMC 3024668. PMID 21199946.
- "Correction for Traverse and Ochman, Conserved rates and patterns of transcription errors across bacterial growth states and lifestyles". Proceedings of the National Academy of Sciences. 113 (29): E4257–E4258. 19 July 2016. doi:10.1073/pnas.1609677113. PMC 4961203. PMID 27402746.
- Xiong, Kun; McEntee, Jay P.; Porfirio, David J.; Masel, Joanna (January 2017). "Drift Barriers to Quality Control When Genes Are Expressed at Different Levels". Genetics. 205 (1): 397–407. doi:10.1534/genetics.116.192567. PMC 5223517. PMID 27838629.
- Meer, K M; Nelson, P G; Xiong, K; Masel, J (16 December 2019). "High transcriptional error rates vary as a function of gene expression level". Genome Biology and Evolution. 12: 3754–3761. doi:10.1093/gbe/evz275. PMC 6988749. PMID 31841128.
See also
External links
- The Nearly Neutral Theory of Molecular Evolution - Perspectives on Molecular Evolution