Nick (DNA)
A nick is a discontinuity in a double stranded DNA molecule where there is no phosphodiester bond between adjacent nucleotides of one strand typically through damage or enzyme action. Nicks allow DNA strands to untwist during replication, and are also thought to play a role in the DNA mismatch repair mechanisms that fix errors on both the leading and lagging daughter strands.[1]
Formation of nicks
The diagram shows the effects of nicks on intersecting DNA in a twisted plasmid. Nicking can be used to dissipate the energy held up by intersecting states. The nicks allow the DNA to take on a circular shape.[2]
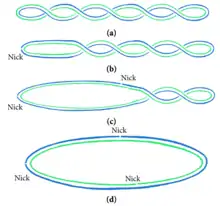
Nicked DNA can be the result of DNA damage or purposeful, regulated biomolecular reactions carried out in the cell. During processing, DNA can be nicked by physical shearing, over-drying or enzymes. Excessive rough handling in pipetting or vortexing creates physical stress that can lead to breaks and nicks in DNA. Overdrying of DNA can also break the phosphodiester bond in DNA and result in nicks. Nicking endonuclease enzymes can assist with this process. A single-stranded break (nick) in DNA can be formed by the hydrolysis and subsequent removal of a phosphate group within the helical backbone. This leads to a different DNA conformation, where a hydrogen bond forms in place of the missing piece of the DNA backbone in order to preserve the structure.[3]
Repair of nicks
Ligases are versatile and ubiquitous enzymes that join the 3’ hydroxyl and 5’ phosphate ends to form a phosphodiester bond, making them essential in nicked DNA repair, and ultimately genome fidelity. This biological role has also been extremely valuable in sealing the sticky ends of plasmids in molecular cloning. Their importance is attested by the fact most organisms have multiple ligases dedicated to specific pathways of repairing DNA. In eubacteria these ligases are powered by NAD+ rather than ATP.[4] Each nick site requires 1 ATP or 1 NAD+ to power the ligase repair.[4]
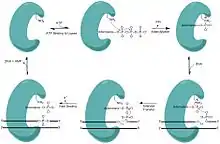
In order to join these fragments, the ligase progresses through three steps:
- Addition of an adenosine monophosphate (AMP) group to the enzyme, referred to as adenylylation,
- Adenosine monophosphate transfer to the DNA and
- Nick sealing, or phosphodiester bond formation.[5][6]
One particular example of a ligase catalyzing nick closure is the E. coli NAD+ dependent DNA ligase, LigA. LigA is a relevant example as it is structurally similar to a clade of enzymes found across all types of bacteria.[7]
Ligases have a metal binding site which is capable of recognizing nicks in DNA. The ligase forms a DNA-adenylate complex, assisting recognition.[8] With human DNA ligase, this forms a crystallized complex. The complex, which has a DNA–adenylate intermediate, allows DNA ligase I to institute a conformational change in the DNA for the isolation and subsequent repair of the DNA nick.[9]
Biological implications
Role in mismatch repair
Single-stranded nicks act as recognizable markers to help the repair machinery distinguish the newly synthesized strand (daughter strand) from the template strand (parental strand).[1] DNA mismatch repair (MMR) is an important DNA repair system that helps maintain genome plasticity by correcting mismatches, or non Watson-Crick base pairs in the a DNA duplex.[10] Some sources of mismatched base pairs include replication errors and deamination of 5-methylcytosine DNA to form thymine. MMR in most bacteria and eukaryotes is directed to the erroneous strand of the mismatched duplex through recognition of strand discontinuities, while MMR in E. coli and closely related bacteria is directed to the strand on the basis of the absence of methylation. Nicking endonucleases introduce the strand discontinuities, or DNA nicks, for both respective systems. Mut L homologues from eukaryotes and most bacteria incise the discontinuous strand to introduce the entry or termination point for the excision reaction. Similarly, in E. coli, Mut H nicks the unmethylated strand of the duplex to introduce the entry point of excision.[11] For eukaryotes specifically, the mechanism of DNA replication elongation between the leading and lagging strand differs. On the lagging strand, nicks exist between Okazaki fragments and are easily recognizable by the DNA mismatch repair machinery prior to ligation. Due to the continuous replication that occurs on the leading strand, the mechanism there is slightly more complex. During replication, ribonucleotides are added by replication enzymes and these ribonucleotides are nicked by an enzyme called RNase H2.[1] Together, the presence of a nick and a ribonucleotide make the leading strand easily recognizable to the DNA mismatch repair machinery.
Nick translation is a biological process in which a single-stranded DNA nick serves as the marker for DNA polymerase to excise and replace possibly damaged nucleotides.[3] At the end of the segment that DNA polymerase acts on, DNA ligase must repair the final segment of DNA backbone in order to complete the repair process.[4] In a lab setting, this can be used to introduce fluorescent or other tagged nucleotides by purposefully inducing site-specific, single-stranded nicks in DNA in vitro and then adding the nicked DNA to an environment rich in DNA polymerase and tagged nucleotide. The DNA polymerase then replaces the DNA nucleotides with the tagged ones, starting at the site of the single-stranded nick.
Role in replication and transcription
Nicked DNA plays an important role in many biological functions. For instance, single-stranded nicks in DNA may serve as purposeful biological markers for the enzyme topoisomerase that unwinds packed DNA and is critical to DNA replication and transcription. In these instances, nicked DNA is not the result of unwanted cell damage.[2]
Topoisomerase-1 preferentially acts at nicks in DNA to cleave adjacent to the nick and then winds or unwinds the complex topologies associated with packed DNA. Here, the nick in the DNA serves as a marker for single strand breakage and subsequent unwinding.[12] It is possible that this is not a highly conserved process. Topoisomerase may cause short deletions when it cleaves bonds, because both full-length DNA products and short deletion strands are seen as products of topoisomerase cleavage while inactive mutants only produced full-length DNA strands.[13]
Nicks in DNA also give rise to different structural properties, can be involved in repairing damages caused by ultraviolet radiation, and are used in the primary steps that allow for genetic recombination.[14]
Nick idling is a biological process in which DNA polymerase may slow or stop its activity of adding bases to a new daughter strand during DNA replication at a nick site.[4] This is particularly relevant to Okazaki fragments in lagging strand in double stranded DNA replication because the direction of replication is opposite to the direction of DNA polymerase, therefore nick idling plays a role in stalling the complex as it replicates in the reverse direction in small fragments (Okazaki fragments) and has to stop and reposition itself in between each and every fragment length of DNA.
DNA structure changes when a single-stranded nick is introduced.[14] Stability is decreased as a break in the phosphodiester backbone allows DNA to unwind, as the built up stress from twisting and packing is not being resisted as strongly anymore.[12] Nicked DNA is more susceptible to degradation due to this reduced stability.
In bacteria
The nic site or nick region is found within the origin of transfer (oriT) site and is a key in starting bacterial conjugation. A single strand of DNA, called the T-strand, is cut at nic by an enzyme called relaxase.[15] This single strand is eventually transferred to the recipient cell during the process of bacterial conjugation. Before this cleavage can occur, however, it is necessary for a group of proteins to attach to the oriT site. This group of proteins is called the relaxosome.[15] It is thought that portions of the oriT site are bent in a way that creates interaction between the relaxosome proteins and the nic site.[15]
Cleaving the T-strand involves relaxase cutting a phosphodiester bond at the nic site.[15] The cleaved strand is left with a hydroxyl group at the 3' end, which may allow for the strand to form a circular plasmid after moving into the recipient cell.[16][17]
References
- Williams JS, Kunkel TA. Ribonucleotides in DNA: Origins, repair and consequences. DNA repair. 2014;19:27-37. doi:10.1016/j.dnarep.2014.03.029
- Chao Ji, Lingyun Zhang, and Pengye Wang, "Configuration Transitions of Free Circular DNA System Induced by Nicks," Journal of Nanomaterials, vol. 2015, Article ID 546851, 7 pages, 2015. doi:10.1155/2015/546851
- Aymami, J.; Coll, M.; Marel, G. A. van der; Boom, J. H. van; Wang, A. H.; Rich, A. (1990-04-01). "Molecular structure of nicked DNA: a substrate for DNA repair enzymes". Proceedings of the National Academy of Sciences. 87 (7): 2526–2530. doi:10.1073/pnas.87.7.2526. ISSN 0027-8424. PMC 53722. PMID 2320572.
- Timson, David J; Singleton, Martin R; Wigley, Dale B (2000-08-30). "DNA ligases in the repair and replication of DNA". Mutation Research/DNA Repair. 460 (3–4): 301–318. doi:10.1016/S0921-8777(00)00033-1. PMID 10946235.
- Ellenberger T, Tomkinson AE (2008). "Eukaryotic DNA ligases: structural and functional insights". Annu. Rev. Biochem. 77: 313–38. doi:10.1146/annurev.biochem.77.061306.123941. PMC 2933818. PMID 18518823.
- Sriskanda V, Shuman S (January 1998). "Chlorella virus DNA ligase: nick recognition and mutational analysis". Nucleic Acids Res. 26 (2): 525–31. doi:10.1093/nar/26.2.525. PMC 147278. PMID 9421510.
- Nandakumar, Jayakrishnan; Nair, Pravin A.; Shuman, Stewart (2007-04-27). "Last stop on the road to repair: structure of E. coli DNA ligase bound to nicked DNA-adenylate". Molecular Cell. 26 (2): 257–271. doi:10.1016/j.molcel.2007.02.026. ISSN 1097-2765. PMID 17466627.
- Odell, Mark; Sriskanda, Verl; Shuman, Stewart; Nikolov, Dimitar B. (2000-11-01). "Crystal Structure of Eukaryotic DNA Ligase–Adenylate Illuminates the Mechanism of Nick Sensing and Strand Joining". Molecular Cell. 6 (5): 1183–1193. doi:10.1016/S1097-2765(00)00115-5. PMID 11106756.
- Pascal, John M.; O'Brien, Patrick J.; Tomkinson, Alan E.; Ellenberger, Tom (2004-11-25). "Human DNA ligase I completely encircles and partially unwinds nicked DNA". Nature. 432 (7016): 473–478. doi:10.1038/nature03082. ISSN 1476-4687. PMID 15565146.
- Morris, James (2013). Biology: How Life Works. W. H. Freeman. pp. Chapter 14, "Mutations and DNA Repair". ISBN 978-1-319-05691-9.
- Fukui, Kenji (2010-07-27). "DNA Mismatch Repair in Eukaryotes and Bacteria". Journal of Nucleic Acids. 2010: 1–16. doi:10.4061/2010/260512. ISSN 2090-0201. PMC 2915661. PMID 20725617.
- Huang, Shar-Yin Naomi; Ghosh, Sanchari; Pommier, Yves (2015-05-29). "Topoisomerase I alone is sufficient to produce short DNA deletions and can also reverse nicks at ribonucleotide sites". The Journal of Biological Chemistry. 290 (22): 14068–14076. doi:10.1074/jbc.M115.653345. ISSN 1083-351X. PMC 4447978. PMID 25887397.
- Racko, Dusan; Benedetti, Fabrizio; Dorier, Julien; Burnier, Yannis; Stasiak, Andrzej (2015-07-06). "Generation of supercoils in nicked and gapped DNA drives DNA unknotting and postreplicative decatenation". Nucleic Acids Research. 43 (15): 7229–7236. doi:10.1093/nar/gkv683. ISSN 0305-1048. PMC 4551925. PMID 26150424.
- Hays, J. B.; Zimm, B. H. (1970-03-14). "Flexibility and stiffness in nicked DNA". Journal of Molecular Biology. 48 (2): 297–317. doi:10.1016/0022-2836(70)90162-2. ISSN 0022-2836. PMID 5448592.
- Lanka Erich, Wilkins Brian M (1995). “DNA Processing Reactions in Bacterial Conjugation.” Annu. Rev. Biochem. 64: 141-69
- Matson S W, Nelson W C, Morton B S (1993). "Characterization of the Reaction Product of the oriT Nicking Reaction Catalyzed by Escherichia coli DNA helicase I." Journal of Bacteriology. 175(9): 2599-2606.
- Grohmann E, Muth G, Espinosa M (2003). "Conjugative Plasmid Transfer in Gram-positive Bacteria." Microbiol Mol Biol Rev. 67(2):277-301.