Nidovirales
Nidovirales is an order of enveloped, positive-strand RNA viruses which infect vertebrates and invertebrates. Host organisms include mammals, birds, reptiles, amphibians, fish, arthropods, molluscs, and helminths.[1] The order includes the families Coronaviridae, Arteriviridae, Roniviridae, and Mesoniviridae.[2]
| Nidovirales | |
|---|---|
 | |
| Life cycle of nidoviruses | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Kingdom: | Orthornavirae |
| Phylum: | Pisuviricota |
| Class: | Pisoniviricetes |
| Order: | Nidovirales |
Member viruses have a viral envelope and a positive-sense, single-stranded RNA genome which is capped and polyadenylated.[3] Nidoviruses are named for the Latin nidus, meaning nest, as all viruses in this order produce a 3' co-terminal nested set of subgenomic mRNAs during infection.[4]
Virology
The order Nidovirales consists of viruses which have positive-sense, single-stranded RNA genomes. The viruses belong to Group IV of the Baltimore classification system. As positive-sense genomes, the viruses can use some host cell proteins during replication and gene expression which occurs in the cytoplasm of the host cell.
This group of viruses expresses structural proteins separately from the nonstructural ones. The structural proteins are encoded at the 3’ region of the genome and are expressed from a set of subgenomic mRNAs. These viruses encode one main proteinase and between one and three accessory proteinases which are mainly involved in expressing the replicase gene. These proteinases are also responsible for activating or inactivating specific proteins at the correct time in the virus life cycle, ensuring replication occurs at the right time.
Many proteins have been identified on the genomes of Nidovirales, but their function has not yet been determined. Other enzymes that may be present in the genome include papain-like proteases, ADP-ribose/poly(ADP-ribose)-binding or ADP-ribose 1''-phosphate phosphatase activities and cyclic nucleotide phosphodiesterase.
Most, but not all, nidovirus subgenomic RNAs contain a 5′ leader sequence derived from the 5′ end of the genomic RNA.
The frameshift that generates ORF1b frameshift occurs at a UUUAAAC heptanucleotide 'slippery' sequence located upstream of the ORF1a stop codon and a putative RNA pseudoknot structure.
Genome
This order of viruses can be distinguished from other RNA viruses by a constellation of seven conserved domains—5'-TM2-3CLpro-TM3-RdRp-Zm-HEL1-NendoU-3'—with the first three being encoded in ORF1a and the remaining four in ORF1b. TM2 and TM3 and transmembrane domains; RdRp is the RNA polymerase; Zm is a Zn-cluster binding domain fused with a helicase (HEL1); 3CLpro is a 3C-like protease; and NendoU is an uridylate-specific endonuclease. The 3CLpro has a catalytic His-Cys dyad, and is also known as the SARS coronavirus main proteinase (Mpro).
Taxonomy
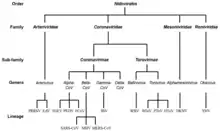
The order Nidovirales can be divided into two clades depending on the size of the genome: those with large genomes (26.3–31.7 kilobases) which included the Coronaviridae and Roniviridae (the large nidoviruses) and those with small genomes (the small nidoviruses)—a clade that includes the distantly related Arteriviridae (12.7–15.7 kb). The large nidoviruses encode both a 2'-O-methyltransferase and a 3'–5' exoribonuclease (ExoN)—the latter being very unusual for an RNA virus. They also encode a superfamily 1 helicase, uridylate-specific endonuclease (an enzyme unique to nidoviruses) and several proteases.
The following suborders and families are recognized (-virineae denotes suborders and -viridae denotes families):[5]
- Abnidovirineae
- Arnidovirineae
- Arteriviridae
- Cremegaviridae
- Gresnaviridae
- Olifoviridae
- Cornidovirineae
- Mesnidovirineae
- Medioniviridae
- Mesoniviridae
- Monidovirineae
- Nanidovirinae
- Nanghoshaviridae
- Nanhypoviridae
- Ronidovirineae
- Euroniviridae
- Roniviridae
- Tornidovirineae
Additions
The virus which has the largest known nonsegmented RNA genome of 41.1kb, the planarian secretory cell nidovirus (PSCNV), is of the order Nidovirales.[6] Its host is the planarian flatworm.[7]
See also
References
- Ogando, Natacha S.; Ferron, Francois; Decroly, Etienne; Canard, Bruno; Posthuma, Clara C.; Snijder, Eric J. (2019). "The Curious Case of the Nidovirus Exoribonuclease: Its Role in RNA Synthesis and Replication Fidelity". Frontiers in Microbiology. 10: 1813. doi:10.3389/fmicb.2019.01813. ISSN 1664-302X. PMC 6693484. PMID 31440227.
- "International Committee on Taxonomy of Viruses (ICTV)". talk.ictvonline.org. Retrieved 2020-06-08.
- King, Andrew M. Q.; Adams, Michael J.; Carstens, Eric B.; Lefkowitz, Elliot J., eds. (2012-01-01), "Order - Nidovirales", Virus Taxonomy, Elsevier, pp. 784–794, ISBN 978-0-12-384684-6, retrieved 2020-06-08
- Antoine A.F. de Vries, Marian C. Horzinek, Peter J. M. Rottier, Raoul J. de Groot (1997). "The Genome Organization of the Nidovirales: Similarities and Differences between Arteri-, Toro-, and Coronaviruses". Seminars in Virology. 8 (1): 33–47. CiteSeerX 10.1.1.462.1825. doi:10.1006/smvy.1997.0104. S2CID 85383257.CS1 maint: uses authors parameter (link)
- "Virus Taxonomy: 2019 Release". talk.ictvonline.org. International Committee on Taxonomy of Viruses. Retrieved 30 April 2020.
- "Taxonomy browser (Planidovirus 1)". www.ncbi.nlm.nih.gov. Retrieved 2020-06-08.
- Saberi, Amir; Gulyaeva, Anastasia A.; Brubacher, John L.; Newmark, Phillip A.; Gorbalenya, Alexander E. (2018-11-01). "A planarian nidovirus expands the limits of RNA genome size". PLOS Pathogens. 14 (11): e1007314. doi:10.1371/journal.ppat.1007314. ISSN 1553-7374. PMC 6211748. PMID 30383829.
- Ziebuhr, J; Snijder, EJ; Gorbalenya, AE (April 2000). "Virus-encoded proteinases and proteolytic processing in the Nidovirales" (PDF). The Journal of General Virology. 81 (Pt 4): 853–79. doi:10.1099/0022-1317-81-4-853. PMID 10725411. Archived from the original (PDF) on 2008-12-16. Retrieved 2007-02-23.
External links
- Nidovirales—ICTVdB—The Universal Virus Database, version 4.
- Nidovirales at the US National Library of Medicine Medical Subject Headings (MeSH)
- NIH/MeSH
- "The Nidoviruses" ISBN 0306466341
