PIKFYVE
PIKfyve, a FYVE finger-containing phosphoinositide kinase, is an enzyme that in humans is encoded by the PIKFYVE gene.[5][6]
Function
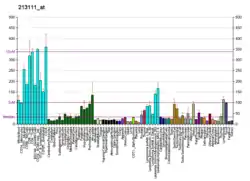
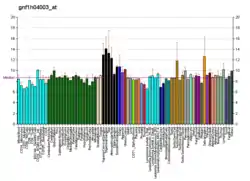
The principal enzymatic activity of PIKfyve is to phosphorylate PtdIns3P to PtdIns(3,5)P2. PIKfyve activity is responsible for the production of both PtdIns(3,5)P2 and phosphatidylinositol 5-phosphate (PtdIns5P).[7][8][9][10] PIKfyve is a large protein, containing a number of functional domains and expressed in several spliced forms. The reported full-length mouse and human cDNA clones encode proteins of 2052 and 2098 amino acid residues, respectively.[6][11][12][13] By directly binding membrane PtdIns(3)P,[14] the FYVE finger domain of PIKfyve is essential in localizing the protein to the cytosolic leaflet of endosomes.[6][14] Impaired PIKfyve enzymatic activity by dominant-interfering mutants, siRNA- mediated ablation or pharmacological inhibition causes lysosome enlargement and cytoplasmic vacuolation due to impaired PtdIns(3,5)P2 synthesis and impaired lysosome fission process and homeostasis.[15] Thus, via PtdIns(3,5)P2 production, PIKfyve participates in several aspects of vesicular dynamics,[16][17] thereby affecting a number of trafficking pathways that emanate from or traverse the endosomal system en route to the trans-Golgi network or later compartments along the endocytic pathway.[18][19][20][21][22][23]
Medical significance
PIKfyve mutations affecting one of the two PIKFYVE alleles are found in 8 out of 10 families with Francois-Neetens corneal fleck dystrophy.[24] Disruption of both PIKFYVE alleles in the mouse is lethal at the stage of pre-implantation embryo.[25] PIKfyve’s role in pathogen invasion is deduced by evidence from cell studies implicating PIKfyve activity in HIV and Salmonella replication.[21][26][27] A link of PIKfyve with type 2 diabetes is inferred by the observations that PIKfyve perturbation inhibits insulin-regulated glucose uptake.[28][29] Concordantly, mice with selective Pikfyve gene disruption in skeletal muscle, the tissue mainly responsible for the decrease of postprandial blood sugar, exhibit systemic insulin resistance; glucose intolerance; hyperinsulinemia; and increased adiposity, i.e. symptoms, typical for human prediabetes.[30]
PIKfyve inhibitors as potential therapeutics in Cancer
Several small molecule PIKfyve inhibitors have shown promise as cancer therapeutics in preclinical studies due to selective toxicity in non-Hodgkin lymphoma B cells [31] or in U-251 glioblastoma cells. [32] PIKfyve inhibitors cause cell death also in A-375 melanoma cells, which depend on autophagy for growth and proliferation, due to impaired lysosome homeostasis. [33] The potential therapeutic use of PIKfyve inhibitors awaits clinical trials.
Interactions
PIKfyve physically associates with its regulator ArPIKfyve, a protein encoded by the human gene VAC14, and the Sac1 domain-containing PtdIns(3,5)P2 5-phosphatase Sac3, encoded by FIG4, to form a stable ternary heterooligomeric complex that is scaffolded by ArPIKfyve homooligomeric interactions. The presence of two enzymes with opposing activities for PtdIns(3,5)P2 synthesis and turnover in a single complex indicates the requirement for a tight control of PtdIns(3,5)P2 levels.[17][34][35] PIKfyve also interacts with the Rab9 effector RABEPK and the kinesin adaptor JLP, encoded by SPAG9.[19][23] These interactions link PIKfyve to microtubule-based endosome to trans-Golgi network traffic. Under sustained activation of glutamate receptors PIKfyve binds to and facilitates the lysosomal degradation of Cav1.2, voltage-dependent calcium channel type 1.2, thereby protecting the neurons from excitotoxicity.[36] PIKfyve negatively regulates Ca2+-dependent exocytosis in neuroendocrine cells without affecting voltage-gated calcium channels.[37]
Evolutionary biology
PIKFYVE belongs to a large family of evolutionarily-conserved lipid kinases. Single copy genes, encoding similarly-structured FYVE-domain–containing phosphoinositide kinases exist in most genomes from yeast to man. The plant A. thaliana has several copies of the enzyme. Higher eukaryotes (after D. melanogaster), acquire an additional DEP domain. The S. cerevisiae enzyme Fab1p is required for PtdIns(3,5)P2 synthesis under basal conditions and in response to hyperosmotic shock. PtdIns5P, made by PIKfyve kinase activity in mammalian cells, is not detected in budding yeast.[38] Yeast Fab1p associates with Vac14p (the ortholog of human ArPIKfyve) and Fig4p (the ortholog of Sac3).[39] The yeast Fab1 complex also includes Vac7p and probably Atg18p, proteins that are not detected in the mammalian PIKfyve complex.[40] S. cerevisiae could survive without Fab1.[41] In contrast, the knockout of the FYVE domain-containing enzymes in A. thaliana, D. melanogaster, C. elegans and M. musculus leads to embryonic lethality indicating that the FYVE-domain–containing phosphoinositide kinases have become essential in embryonic development of multicellular organisms.[25][42][43][44] Thus, in evolution, the FYVE-domain-containing phosphoinositide kinases retain several aspects of the structural organization, enzyme activity and protein interactions from budding yeast. In higher eukaryotes, the enzymes acquire one additional domain, a role in the production of PtdIns5P, a new set of interacting proteins and become essential in embryonic development.
References
- GRCh38: Ensembl release 89: ENSG00000115020 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000025949 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Entrez Gene: Phosphoinositide kinase, FYVE finger containing".
- Shisheva A, Sbrissa D, Ikonomov O (January 1999). "Cloning, characterization, and expression of a novel Zn2+-binding FYVE finger-containing phosphoinositide kinase in insulin-sensitive cells". Molecular and Cellular Biology. 19 (1): 623–34. doi:10.1128/MCB.19.1.623. PMC 83920. PMID 9858586.
- Shisheva A (2001). "PIKfyve: the road to PtdIns 5-P and PtdIns 3,5-P(2)". Cell Biology International. 25 (12): 1201–6. doi:10.1006/cbir.2001.0803. PMID 11748912. S2CID 29411107.
- Sbrissa D, Ikonomov OC, Deeb R, Shisheva A (December 2002). "Phosphatidylinositol 5-phosphate biosynthesis is linked to PIKfyve and is involved in osmotic response pathway in mammalian cells". The Journal of Biological Chemistry. 277 (49): 47276–84. doi:10.1074/jbc.M207576200. PMID 12270933.
- Sbrissa D, Ikonomov OC, Filios C, Delvecchio K, Shisheva A (August 2012). "Functional dissociation between PIKfyve-synthesized PtdIns5P and PtdIns(3,5)P2 by means of the PIKfyve inhibitor YM201636". American Journal of Physiology. Cell Physiology. 303 (4): C436-46. doi:10.1152/ajpcell.00105.2012. PMC 3422984. PMID 22621786.
- Zolov SN, Bridges D, Zhang Y, Lee WW, Riehle E, Verma R, et al. (October 2012). "In vivo, Pikfyve generates PI(3,5)P2, which serves as both a signaling lipid and the major precursor for PI5P". Proceedings of the National Academy of Sciences of the United States of America. 109 (43): 17472–7. doi:10.1073/pnas.1203106109. PMC 3491506. PMID 23047693.
- Sbrissa D, Ikonomov OC, Shisheva A (July 1999). "PIKfyve, a mammalian ortholog of yeast Fab1p lipid kinase, synthesizes 5-phosphoinositides. Effect of insulin". The Journal of Biological Chemistry. 274 (31): 21589–97. doi:10.1074/jbc.274.31.21589. PMID 10419465.
- Sbrissa D, Ikonomov OC, Deeb R, Shisheva A (December 2002). "Phosphatidylinositol 5-phosphate biosynthesis is linked to PIKfyve and is involved in osmotic response pathway in mammalian cells". The Journal of Biological Chemistry. 277 (49): 47276–84. doi:10.1074/jbc.M207576200. PMID 12270933.
- Cabezas A, Pattni K, Stenmark H (April 2006). "Cloning and subcellular localization of a human phosphatidylinositol 3-phosphate 5-kinase, PIKfyve/Fab1". Gene. 371 (1): 34–41. doi:10.1016/j.gene.2005.11.009. PMID 16448788.
- Sbrissa D, Ikonomov OC, Shisheva A (February 2002). "Phosphatidylinositol 3-phosphate-interacting domains in PIKfyve. Binding specificity and role in PIKfyve. Endomenbrane localization". The Journal of Biological Chemistry. 277 (8): 6073–9. doi:10.1074/jbc.M110194200. PMID 11706043.
- Sharma G, Guardia CM, Roy A, Vassilev A, Saric A, Griner LN, et al. (February 2019). "A family of PIKFYVE inhibitors with therapeutic potential against autophagy-dependent cancer cells disrupt multiple events in lysosome homeostasis". Autophagy. 15 (10): 1694–1718. doi:10.1080/15548627.2019.1586257. PMC 6735543. PMID 30806145.
- Ikonomov OC, Sbrissa D, Shisheva A (August 2006). "Localized PtdIns 3,5-P2 synthesis to regulate early endosome dynamics and fusion". American Journal of Physiology. Cell Physiology. 291 (2): C393-404. doi:10.1152/ajpcell.00019.2006. PMID 16510848.
- Sbrissa D, Ikonomov OC, Fu Z, Ijuin T, Gruenberg J, Takenawa T, Shisheva A (August 2007). "Core protein machinery for mammalian phosphatidylinositol 3,5-bisphosphate synthesis and turnover that regulates the progression of endosomal transport. Novel Sac phosphatase joins the ArPIKfyve-PIKfyve complex". The Journal of Biological Chemistry. 282 (33): 23878–91. doi:10.1074/jbc.M611678200. PMID 17556371.
- Ikonomov OC, Sbrissa D, Shisheva A (July 2001). "Mammalian cell morphology and endocytic membrane homeostasis require enzymatically active phosphoinositide 5-kinase PIKfyve". The Journal of Biological Chemistry. 276 (28): 26141–7. doi:10.1074/jbc.M101722200. PMID 11285266.
- Ikonomov OC, Sbrissa D, Mlak K, Deeb R, Fligger J, Soans A, et al. (December 2003). "Active PIKfyve associates with and promotes the membrane attachment of the late endosome-to-trans-Golgi network transport factor Rab9 effector p40". The Journal of Biological Chemistry. 278 (51): 50863–71. doi:10.1074/jbc.M307260200. PMID 14530284.
- Rutherford AC, Traer C, Wassmer T, Pattni K, Bujny MV, Carlton JG, et al. (October 2006). "The mammalian phosphatidylinositol 3-phosphate 5-kinase (PIKfyve) regulates endosome-to-TGN retrograde transport". Journal of Cell Science. 119 (Pt 19): 3944–57. doi:10.1242/jcs.03153. PMC 1904490. PMID 16954148.
- Jefferies HB, Cooke FT, Jat P, Boucheron C, Koizumi T, Hayakawa M, et al. (February 2008). "A selective PIKfyve inhibitor blocks PtdIns(3,5)P(2) production and disrupts endomembrane transport and retroviral budding". EMBO Reports. 9 (2): 164–70. doi:10.1038/sj.embor.7401155. PMC 2246419. PMID 18188180.
- Shisheva A (June 2008). "PIKfyve: Partners, significance, debates and paradoxes". Cell Biology International. 32 (6): 591–604. doi:10.1016/j.cellbi.2008.01.006. PMC 2491398. PMID 18304842.
- Ikonomov OC, Fligger J, Sbrissa D, Dondapati R, Mlak K, Deeb R, Shisheva A (February 2009). "Kinesin adapter JLP links PIKfyve to microtubule-based endosome-to-trans-Golgi network traffic of furin". The Journal of Biological Chemistry. 284 (6): 3750–61. doi:10.1074/jbc.M806539200. PMC 2635046. PMID 19056739.
- Li S, Tiab L, Jiao X, Munier FL, Zografos L, Frueh BE, et al. (July 2005). "Mutations in PIP5K3 are associated with François-Neetens mouchetée fleck corneal dystrophy". American Journal of Human Genetics. 77 (1): 54–63. doi:10.1086/431346. PMC 1226194. PMID 15902656.
- Ikonomov OC, Sbrissa D, Delvecchio K, Xie Y, Jin JP, Rappolee D, Shisheva A (April 2011). "The phosphoinositide kinase PIKfyve is vital in early embryonic development: preimplantation lethality of PIKfyve-/- embryos but normality of PIKfyve+/- mice". The Journal of Biological Chemistry. 286 (15): 13404–13. doi:10.1074/jbc.M111.222364. PMC 3075686. PMID 21349843.
- Murray JL, Mavrakis M, McDonald NJ, Yilla M, Sheng J, Bellini WJ, et al. (September 2005). "Rab9 GTPase is required for replication of human immunodeficiency virus type 1, filoviruses, and measles virus". Journal of Virology. 79 (18): 11742–51. doi:10.1128/JVI.79.18.11742-11751.2005. PMC 1212642. PMID 16140752.
- Kerr MC, Wang JT, Castro NA, Hamilton NA, Town L, Brown DL, et al. (April 2010). "Inhibition of the PtdIns(5) kinase PIKfyve disrupts intracellular replication of Salmonella". The EMBO Journal. 29 (8): 1331–47. doi:10.1038/emboj.2010.28. PMC 2868569. PMID 20300065.
- Ikonomov OC, Sbrissa D, Mlak K, Shisheva A (December 2002). "Requirement for PIKfyve enzymatic activity in acute and long-term insulin cellular effects". Endocrinology. 143 (12): 4742–54. doi:10.1210/en.2002-220615. PMID 12446602.
- Ikonomov OC, Sbrissa D, Dondapati R, Shisheva A (July 2007). "ArPIKfyve-PIKfyve interaction and role in insulin-regulated GLUT4 translocation and glucose transport in 3T3-L1 adipocytes". Experimental Cell Research. 313 (11): 2404–16. doi:10.1016/j.yexcr.2007.03.024. PMC 2475679. PMID 17475247.
- Ikonomov, O. C.; Sbrissa, D.; Delvecchio, K.; Feng, H. Z.; Cartee, G. D.; Jin, J. P.; Shisheva, A. (2013). "Muscle-specific Pikfyve gene disruption causes glucose intolerance, insulin resistance, adiposity, and hyperinsulinemia but not muscle fiber-type switching". American Journal of Physiology. Endocrinology and Metabolism. 305 (1): E119-31. doi:10.1152/ajpendo.00030.2013. PMC 3725567. PMID 23673157.
- Gayle, S; Landrette, S; Beeharry, N; Conrad, C; Hernandez, M; Beckett, P; Ferguson, SM; Mendelkern, T; Zheng, M; Xu, T; Rothberg, J; Lichenstein, H (2017). "Identification of apilimod as a first-in-class PIKfyve kinase inhibitor for treatment of B-cell non-Hodgkin lymphoma". Blood. 129 (13): 1768–1778. doi:10.1182/blood-2016-09-736892. PMC 5766845. PMID 28104689.
- Li, Z; Mbah, NE; Overmeyer, JH; Sarver, JG; George, S; Trabbic, CJ; Erhardt, PW; Maltese, WA (2019). "The JNK signaling pathway plays a key role in methuosis (non-apoptotic cell death) induced by MOMIPP in glioblastoma". BMC Cancer. 19 (1): 77. doi:10.1186/s12885-019-5288-y. PMC 6335761. PMID 30651087.
- Sharma G, Guardia CM, Roy A, Vassilev A, Saric A, Griner LN, et al. (February 2019). "A family of PIKFYVE inhibitors with therapeutic potential against autophagy-dependent cancer cells disrupt multiple events in lysosome homeostasis". Autophagy. 15 (10): 1694–1718. doi:10.1080/15548627.2019.1586257. PMC 6735543. PMID 30806145.
- Sbrissa D, Ikonomov OC, Fenner H, Shisheva A (December 2008). "ArPIKfyve homomeric and heteromeric interactions scaffold PIKfyve and Sac3 in a complex to promote PIKfyve activity and functionality". Journal of Molecular Biology. 384 (4): 766–79. doi:10.1016/j.jmb.2008.10.009. PMC 2756758. PMID 18950639.
- Ikonomov OC, Sbrissa D, Fenner H, Shisheva A (December 2009). "PIKfyve-ArPIKfyve-Sac3 core complex: contact sites and their consequence for Sac3 phosphatase activity and endocytic membrane homeostasis". The Journal of Biological Chemistry. 284 (51): 35794–806. doi:10.1074/jbc.M109.037515. PMC 2791009. PMID 19840946.
- Tsuruta F, Green EM, Rousset M, Dolmetsch RE (October 2009). "PIKfyve regulates CaV1.2 degradation and prevents excitotoxic cell death". The Journal of Cell Biology. 187 (2): 279–94. doi:10.1083/jcb.200903028. PMC 2768838. PMID 19841139.
- Osborne SL, Wen PJ, Boucheron C, Nguyen HN, Hayakawa M, Kaizawa H, et al. (February 2008). "PIKfyve negatively regulates exocytosis in neurosecretory cells". The Journal of Biological Chemistry. 283 (5): 2804–13. doi:10.1074/jbc.M704856200. PMID 18039667.
- Michell RH, Heath VL, Lemmon MA, Dove SK (January 2006). "Phosphatidylinositol 3,5-bisphosphate: metabolism and cellular functions". Trends in Biochemical Sciences. 31 (1): 52–63. doi:10.1016/j.tibs.2005.11.013. PMID 16364647.
- Botelho RJ, Efe JA, Teis D, Emr SD (October 2008). "Assembly of a Fab1 phosphoinositide kinase signaling complex requires the Fig4 phosphoinositide phosphatase". Molecular Biology of the Cell. 19 (10): 4273–86. doi:10.1091/mbc.E08-04-0405. PMC 2555960. PMID 18653468.
- Jin N, Chow CY, Liu L, Zolov SN, Bronson R, Davisson M, et al. (December 2008). "VAC14 nucleates a protein complex essential for the acute interconversion of PI3P and PI(3,5)P(2) in yeast and mouse". The EMBO Journal. 27 (24): 3221–34. doi:10.1038/emboj.2008.248. PMC 2600653. PMID 19037259.
- Yamamoto A, DeWald DB, Boronenkov IV, Anderson RA, Emr SD, Koshland D (May 1995). "Novel PI(4)P 5-kinase homologue, Fab1p, essential for normal vacuole function and morphology in yeast". Molecular Biology of the Cell. 6 (5): 525–39. doi:10.1091/mbc.6.5.525. PMC 301213. PMID 7663021.
- Rusten TE, Rodahl LM, Pattni K, Englund C, Samakovlis C, Dove S, et al. (September 2006). "Fab1 phosphatidylinositol 3-phosphate 5-kinase controls trafficking but not silencing of endocytosed receptors". Molecular Biology of the Cell. 17 (9): 3989–4001. doi:10.1091/mbc.E06-03-0239. PMC 1556381. PMID 16837550.
- Nicot AS, Fares H, Payrastre B, Chisholm AD, Labouesse M, Laporte J (July 2006). "The phosphoinositide kinase PIKfyve/Fab1p regulates terminal lysosome maturation in Caenorhabditis elegans". Molecular Biology of the Cell. 17 (7): 3062–74. doi:10.1091/mbc.E05-12-1120. PMC 1483040. PMID 16801682.
- Whitley P, Hinz S, Doughty J (December 2009). "Arabidopsis FAB1/PIKfyve proteins are essential for development of viable pollen". Plant Physiology. 151 (4): 1812–22. doi:10.1104/pp.109.146159. PMC 2785992. PMID 19846542.
Further reading
- Nagase T, Ishikawa K, Suyama M, Kikuno R, Hirosawa M, Miyajima N, et al. (February 1999). "Prediction of the coding sequences of unidentified human genes. XIII. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro". DNA Research. 6 (1): 63–70. doi:10.1093/dnares/6.1.63. PMID 10231032.
- Jiao X, Munier FL, Schorderet DF, Zografos L, Smith J, Rubin B, Hejtmancik JF (May 2003). "Genetic linkage of Francois-Neetens fleck (mouchetée) corneal dystrophy to chromosome 2q35". Human Genetics. 112 (5–6): 593–9. doi:10.1007/s00439-002-0905-1. PMID 12607114. S2CID 1338901.
- Ikonomov OC, Sbrissa D, Foti M, Carpentier JL, Shisheva A (November 2003). "PIKfyve controls fluid phase endocytosis but not recycling/degradation of endocytosed receptors or sorting of procathepsin D by regulating multivesicular body morphogenesis". Molecular Biology of the Cell. 14 (11): 4581–91. doi:10.1091/mbc.E03-04-0222. PMC 266774. PMID 14551253.
- Brill LM, Salomon AR, Ficarro SB, Mukherji M, Stettler-Gill M, Peters EC (May 2004). "Robust phosphoproteomic profiling of tyrosine phosphorylation sites from human T cells using immobilized metal affinity chromatography and tandem mass spectrometry". Analytical Chemistry. 76 (10): 2763–72. doi:10.1021/ac035352d. PMID 15144186.
- Sbrissa D, Ikonomov OC, Shisheva A (February 2002). "Phosphatidylinositol 3-phosphate-interacting domains in PIKfyve. Binding specificity and role in PIKfyve. Endomenbrane localization". The Journal of Biological Chemistry. 277 (8): 6073–9. doi:10.1074/jbc.M110194200. PMID 11706043.
- Sbrissa D, Ikonomov OC, Strakova J, Dondapati R, Mlak K, Deeb R, et al. (December 2004). "A mammalian ortholog of Saccharomyces cerevisiae Vac14 that associates with and up-regulates PIKfyve phosphoinositide 5-kinase activity". Molecular and Cellular Biology. 24 (23): 10437–47. doi:10.1128/MCB.24.23.10437-10447.2004. PMC 529046. PMID 15542851.
- Rush J, Moritz A, Lee KA, Guo A, Goss VL, Spek EJ, et al. (January 2005). "Immunoaffinity profiling of tyrosine phosphorylation in cancer cells". Nature Biotechnology. 23 (1): 94–101. doi:10.1038/nbt1046. PMID 15592455. S2CID 7200157.
- Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M (November 2006). "Global, in vivo, and site-specific phosphorylation dynamics in signaling networks". Cell. 127 (3): 635–48. doi:10.1016/j.cell.2006.09.026. PMID 17081983. S2CID 7827573.