Phosphoglucomutase
Phosphoglucomutase (EC 5.4.2.2) is an enzyme that transfers a phosphate group on an α-D-glucose monomer from the 1 to the 6 position in the forward direction or the 6 to the 1 position in the reverse direction.
| Phosphoglucomutase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 | |||||||||
| Identifiers | |||||||||
| EC number | 5.4.2.2 | ||||||||
| CAS number | 9001-81-4 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| |||||||||
More precisely, it facilitates the interconversion of glucose 1-phosphate and glucose 6-phosphate.
Biological Function
Role in glycogenolysis
After glycogen phosphorylase catalyzes the phosphorolytic cleavage of a glucosyl residue from the glycogen polymer, the freed glucose has a phosphate group on its 1-carbon. This glucose 1-phosphate molecule is not itself a useful metabolic intermediate, but phosphoglucomutase catalyzes the conversion of this glucose 1-phosphate to glucose 6-phosphate (see below for the mechanism of this reaction).
Glucose 6-phosphate’s metabolic fate depends on the needs of the cell at the time it is generated. If the cell is low on energy, then glucose 6-phosphate will travel down the glycolytic pathway, eventually yielding two molecules of adenosine triphosphate. If the cell is in need of biosynthetic intermediates, glucose 6-phosphate will enter the pentose phosphate pathway, where it will undergo a series of reactions to yield riboses and/or NADPH, depending on cellular conditions.
If glycogenolysis is taking place in the liver, glucose 6-phosphate can be converted to glucose by the enzyme glucose 6-phosphatase; the glucose produced in the liver is then released to the bloodstream for use in other organs. Muscle cells in contrast do not have the enzyme glucose 6-phosphatase, so they cannot share their glycogen stores with the rest of the body.
Role in glycogenesis
Phosphoglucomutase also acts in the opposite fashion when blood glucose levels are high. In this case, phosphoglucomutase catalyzes the conversion of glucose 6-phosphate (which is easily generated from glucose by the action of hexokinase) to glucose 1-phosphate.
This glucose-1-phosphate can then react with UTP to yield UDP-glucose in a reaction catalyzed by UDP-glucose-pyrophosphorylase. If activated by insulin, glycogen synthase will proceed to clip the glucose from the UDP-glucose complex onto a glycogen polymer.
Reaction mechanism
Phosphoglucomutase affects a phosphoryl group shift by exchanging a phosphoryl group with the substrate.[1] Isotopic labeling experiments have confirmed that this reaction proceeds through a glucose 1,6-bisphosphate intermediate.[2]
The first step in the forward reaction is the transfer of a phosphoryl group from the enzyme to glucose 1-phosphate, forming glucose 1,6-bisphosphate and leaving a dephosphorylated form of the enzyme.[2] The enzyme then undergoes a rapid diffusional reorientation to position the 1-phosphate of the bisphosphate intermediate properly relative to the dephosphorylated enzyme.[3] Substrate-velocity relationships and induced transport tests have revealed that the dephosphorylated enzyme then facilitates the transfer of a phosphoryl group from the glucose-1,6-bisphosphate intermediate to the enzyme, regenerating phosphorylated phosphoglucomutase and yielding glucose 6-phosphate (in the forward direction).[4][5] Later structural studies confirmed that the single site in the enzyme that becomes phosphorylated and dephosphorylated is the oxygen of the active-site serine residue (see diagram below).[6][7] A bivalent metal ion, usually magnesium or cadmium, is required for enzymatic activity and has been shown to complex directly with the phosphoryl group esterified to the active-site serine.[8]
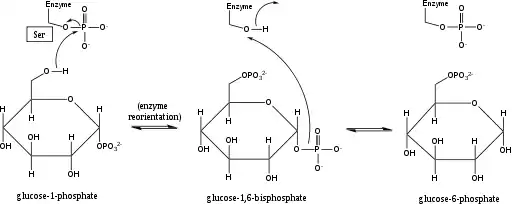
This formation of a glucose 1,6-bisphosphate intermediate is analogous to the interconversion of 2-phosphoglycerate and 3-phosphoglycerate catalyzed by phosphoglycerate mutase, in which 2,3-bisphosphoglycerate is generated as an intermediate.[9]
Structure
While rabbit muscle phosphoglucomutase has served as the prototype for much of the elucidation of this enzyme's structure, newer bacterium-derived crystal structures exhibit many of the same defining characteristics.[10] Each phosphoglucomutase monomer can be divided into four sequence domains, I-IV, based on the enzyme’s default spatial configuration (see image at right).[11]
Each monomer comprises four distinct α/β structural units, each of which contains one of the four strands in each monomer's β-sheet and is made up only of the residues in a given sequence domain (see image at right).[11] The burial of the active site (including Ser-116, the critical residue on the enzyme that is phosphorylated and dephosphorylated) in the hydrophobic interior of the enzyme serves to exclude water from counterproductively hydrolyzing critical phosphoester bonds while still allowing the substrate to access the active site.[12]
Disease relevance
Human muscle contains two phosphoglucomutases with nearly identical catalytic properties, PGM I and PGM II.[13] One or the other of these forms is missing in some humans congenitally.[14]
PGM deficiency is an extremely rare condition that does not have a set of well-characterized physiological symptoms. This condition can be detected by an in vitro study of anaerobic glycolysis which reveals a block in the pathway toward lactic acid production after glucose 1-phosphate but before glucose 6-phosphate.[15]
PGM1 deficiency is known as CDG syndrome type 1t (CDG1T, formerly known as glycogen storage disease type 14 (GSD XIV).[16]
See also
References
- Jagannathan, V; Luck, JM (1949). "Phosphoglucomutase; mechanism of action". Journal of Biological Chemistry. 179 (2): 569–75. doi:10.1016/S0021-9258(19)51252-2. PMID 18149991.
- Najjar, V. A.; Pullman, M. E. (1954). "The Occurrence of a Group Transfer Involving Enzyme (phosphoglucomutase) and Substrate". Science. 119 (3097): 631–4. Bibcode:1954Sci...119..631N. doi:10.1126/science.119.3097.631. PMID 13156640.
- Ray WJ, Peck EJ (1972). "Phosphomutases". The Enzymes. New York: Academic Press.
- Ray, William J.; Roscelli, Gertrude A. (1964). "A Kinetic Study of the Phosphoglucomutase Pathway". Journal of Biological Chemistry. 239 (4): 1228–36. doi:10.1016/S0021-9258(18)91416-X. PMID 14165931.
- Britton, HG; Clarke, JB (1968). "The mechanism of the phosphoglucomutase reaction. Studies on rabbit muscle phosphoglucomutase with flux techniques". Biochemical Journal. 110 (2): 161–80. doi:10.1042/bj1100161. PMC 1187194. PMID 5726186.
- Rayjr, W; Mildvan, A; Grutzner, J (1977). "Phosphorus nuclear magnetic resonance studies of phosphoglucomutase and its metal ion complexes". Archives of Biochemistry and Biophysics. 184 (2): 453–63. doi:10.1016/0003-9861(77)90455-6. PMID 23074.
- Ray Jr, WJ; Hermodson, MA; Puvathingal, JM; Mahoney, WC (1983). "The complete amino acid sequence of rabbit muscle phosphoglucomutase". Journal of Biological Chemistry. 258 (15): 9166–74. doi:10.1016/S0021-9258(17)44646-1. PMID 6223925.
- Rhyu, Gyung Ihm; Ray Jr, William; Markley, John L. (1984). "Enzyme-bound intermediates in the conversion of glucose 1-phosphate to glucose 6-phosphate by phosphoglucomutase. Phosphorus NMR studies". Biochemistry. 23 (2): 252–60. doi:10.1021/bi00297a013. PMID 6230103.
- Sutherland, EW; Cohn, M (1949). "The mechanism of the phosphoglucomutase reaction". Journal of Biological Chemistry. 180 (3): 1285–95. doi:10.1016/S0021-9258(19)51242-X. PMID 18148026.
- Mehra-Chaudhary, Ritcha; Mick, Jacob; Tanner, John J.; Henzl, Michael T.; Beamer, Lesa J. (2011). "Crystal structure of a bacterial phosphoglucomutase, an enzyme involved in the virulence of multiple human pathogens". Proteins: Structure, Function, and Bioinformatics. 79 (4): 1215–29. doi:10.1002/prot.22957. PMC 3066478. PMID 21246636.
- Dai, JB; Liu, Y; Ray Jr, WJ; Konno, M (1992). "The crystal structure of muscle phosphoglucomutase refined at 2.7-angstrom resolution". Journal of Biological Chemistry. 267 (9): 6322–37. doi:10.1016/S0021-9258(18)42699-3. PMID 1532581.
- Ray, William J.; Puvathingal, Joseph M.; Liu, Yiwei (1991). "Formation of substrate and transition-state analog complexes in crystals of phosphoglucomutase after removing the crystallization salt". Biochemistry. 30 (28): 6875–85. doi:10.1021/bi00242a011. PMID 1829964.
- Joshi, JG; Handler, P (1969). "Phosphoglucomutase. VI. Purification and properties of phosphoglucomutases from human muscle". Journal of Biological Chemistry. 244 (12): 3343–51. doi:10.1016/S0021-9258(18)93132-7. PMID 4978319.
- Brown DH (1986). "Glycogen metabolism and glycolysis in muscle". Myology. New York: McGraw-Hill. pp. 673–95.
- Sugie, H; Kobayashi, J; Sugie, Y; Ichimura, M; Miyamoto, R; Ito, T; Shimizu, K; Igarashi, Y (1988). "Infantile muscle glycogen storage disease: phosphoglucomutase deficiency with decreased muscle and serum carnitine levels". Neurology. 38 (4): 602–5. doi:10.1212/WNL.38.4.602. PMID 2965317. S2CID 11491932.
- Orphanet: Glycogen storage disease due to phosphoglucomutase deficiency
External links
- Phosphoglucomutase at the US National Library of Medicine Medical Subject Headings (MeSH)
