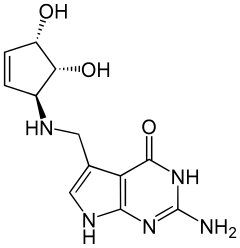
Queuine
Queuine (/kjuːiːn/) (Q) is a hypermodified nucleobase found in the first (or wobble) position of the anticodon of tRNAs specific for Asn, Asp, His, and Tyr, in most eukaryotes and prokaryotes.[1]
 | |
| Names | |
|---|---|
| Other names
7-(((4,5-cis-dihydroxy-2-cyclopenten-1-yl)amino)methyl)-7-deazaguanine | |
| Identifiers | |
3D model (JSmol) |
|
| ChemSpider | |
| MeSH | Queuine |
PubChem CID |
|
CompTox Dashboard (EPA) |
|
| |
| |
| Properties | |
| C12H15N5O3 | |
| Molar mass | 277.284 g·mol−1 |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa). | |
| Infobox references | |
The nucleoside of queuine is queuosine. Queuine is not found in the tRNA of archaea; however, a related 7-deazaguanine derivative, the nucleoside of which is archaeosine, occurs in different tRNA position, the dihydrouridine loop, and in tRNAs with more specificities.
History and naming
In 1967, it was discovered that the four above-mentioned tRNAs contained an as-yet unknown nucleoside, which was designated "Nucleoside Q". This name remained in use throughout much of the work to characterize the compound, after which it was proposed that its common name should be based on the sound of the letter Q—thus producing "queuine" by analogy to guanine and other nucleobases, and "queuosine" by analogy to guanosine and other nucleosides.[2]
Biosynthesis and function
Although queuosine is found in the tRNA of nearly all eukaryotic organisms, it is produced exclusively by bacteria; higher organisms must obtain queuine from the diet or salvage it from symbiotic microbes—a process for which dedicated enzymatic machinery exists.[3] As of 2019, human queuine requirements are not well understood, and the prevalence of queuine deficiency in humans is unknown.[4]
Once salvaged, queuine replaces a guanine base in the anticodon of certain tRNAs, where it appears to play a role in ensuring rapid and accurate recognition of the corresponding mRNAs' codons. In the absence of queuosine modification, translation at Q-decoded codons slows down to the point that many proteins cannot fold properly.[5]
Enzyme research
BH4 is a cofactor for the biopterin-dependent aromatic amino acid hydroxylase enzymes, which catalyze the conversion of phenylalanine to tyrosine, tyrosine to L-DOPA, and tryptophan to 5-HTP, oxidizing BH4 to dihydrobiopterin (BH2) in the process. BH2 must then be converted back to BH4 by the enzyme dihydropteridine reductase before it can be used again. Queuine depletion appears to impair this "recycling" process, resulting in a deficit of BH4 and an excess of BH2, which in turn impairs the activity of the aromatic amino acid hydroxylase enzymes.[6]
References
- Farkas, Walter R. (1983). "Queuine, the Q-Containing tRNAs and the Enzymes Responsible for Their Formation". Nucleosides and Nucleotides. 2: 1–20. doi:10.1080/07328318308078845.
- Nishimura, Susumu; et al. (1983). Cohn, Waldo (ed.). Progress in Nucleic Acid Research and Molecular Biology. 28. 111 Fifth Avenue, New York, New York 10003: Academic Press, Inc. pp. 50–80. ISBN 0-12-540028-4.CS1 maint: location (link)
- Zallot, Rémi; et al. (15 August 2014). "Plant, animal, and fungal micronutrient queuosine is salvaged by members of the DUF2419 protein family". ACS Chemical Biology. 9 (8): 1812–1825. doi:10.1021/cb500278k. PMC 4136680. PMID 24911101.
- Skolnick, Stephen; Greig, Nigel (1 March 2019). "Microbes and monoamines: Potential neuropsychiatric consequences of dysbiosis". Trends in Neurosciences. 42 (3): 151–163. doi:10.1016/j.tins.2018.12.005. PMID 30795845. S2CID 72335336. Retrieved 6 May 2020.
- Tuorto, Francesca; et al. (14 September 2018). "Queuosine-modified tRNAs confer nutritional control of protein translation". EMBO J. 37 (18): e99777. doi:10.15252/embj.201899777. PMC 6138434. PMID 30093495.
- Rakovich, Tatsiana; et al. (12 April 2011). "Queuosine deficiency in eukaryotes compromises tyrosine production through increased tetrahydrobiopterin oxidation". Journal of Biological Chemistry. 286 (22): 19354–19363. doi:10.1074/jbc.M111.219576. PMID 9016755.
External links
- Human Metabolome Database: Queuine (HMDB01495)