Rubredoxin
Rubredoxins are a class of low-molecular-weight iron-containing proteins found in sulfur-metabolizing bacteria and archaea. Sometimes rubredoxins are classified as iron-sulfur proteins; however, in contrast to iron-sulfur proteins, rubredoxins do not contain inorganic sulfide. Like cytochromes, ferredoxins and Rieske proteins, rubredoxins participate in electron transfer in biological systems.
| Rubredoxin | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 rubredoxin domain ii from pseudomonas oleovorans | |||||||||
| Identifiers | |||||||||
| Symbol | Rubredoxin | ||||||||
| Pfam | PF00301 | ||||||||
| Pfam clan | CL0045 | ||||||||
| InterPro | IPR004039 | ||||||||
| PROSITE | PDOC00179 | ||||||||
| SCOP2 | 7rxn / SCOPe / SUPFAM | ||||||||
| |||||||||
Structure
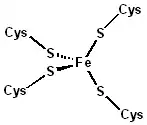
The 3-D structures of a number of rubredoxins have been solved. The fold belongs to the α+β class, with 2 α-helices and 2-3 β-strands. Rubredoxin active site contains an iron ion which is coordinated by the sulfurs of four conserved cysteine residues forming an almost regular tetrahedron. This is sometimes denoted as a [1Fe-0S] or an Fe1S0 system, in analogy to the nomenclature for iron-sulfur proteins. While the vast majority of rubredoxins are soluble, there exists a membrane-bound rubredoxin, referred to as rubredoxin A, in oxygenic photoautotrophs.[1]
Rubredoxins perform one-electron transfer processes. The central iron atom changes between the +2 and +3 oxidation states. In both oxidation states, the metal remains high spin, which helps to minimize structural changes. The reduction potential of a rubredoxin is typically in the range +50 mV to -50 mV.
This iron-sulphur protein is an electron carrier, and it is easy to distinguish its metallic centre changes: the oxidized state is reddish (due to a ligand metal charge transfer), while the reduced state is colourless (because the electron transition has an energy of the infrared level, which is imperceptible to the human eye).
Rubredoxin in some biochemical reactions
- EC 1.14.15.2 camphor 1,2-monooxygenase [(+)-camphor,reduced-rubredoxin:oxygen oxidoreductase (1,2-lactonizing)]
- (+)-bornane-2,5-dione + reduced rubredoxin + O2 = 5-oxo-1,2-campholide + oxidized rubredoxin + H2O
- EC 1.14.15.3 alkane 1-monooxygenase (alkane,reduced-rubredoxin:oxygen 1-oxidoreductase)
- octane + reduced rubredoxin + O2 = 1-octanol + oxidized rubredoxin + H2O
- EC 1.15.1.2 superoxide reductase (rubredoxin:superoxide oxidoreductase)
- reduced rubredoxin + superoxide + 2 H+ = rubredoxin + H2O2
- EC 1.18.1.1 rubredoxin—NAD+ reductase (rubredoxin:NAD+ oxidoreductase)
- reduced rubredoxin + NAD+ = oxidized rubredoxin + NADH + H+
- EC 1.18.1.4 rubredoxin—NAD(P)+ reductase (rubredoxin:NAD(P)+ oxidoreductase)
- reduced rubredoxin + NAD(P)+ = oxidized rubredoxin + NAD(P)H + H+
Electron transfer rate
The electron self-exchange rate is most accurately determined by nuclear magnetic resonance linewidths since the Fe 2+ ions give paramagnetic peak broadening while the Fe+ ion is diamagnetic and therefore causes no broadening.
The electron transfer rate has three parameters it depends on electronic coupling, reorganization energy and free energy of reaction (ΔG°)[2]
Protein mechanism and effects
The amide NH—S-Cys H-bonding lowers the inner sphere reorganization energy giving more rapid electron transfer and the Leu gate stabilizes the Fe 2+ reduced form shifts the redox potential to more positive E0 values. The protein mechanism for the electron transfer of rubredoxin occurs in two steps.[3] The first protein effect, is through the expansion of iron-sulfur bond lengths upon reduction and the shortening of hydrogen bond lengths ensure a better electrostatic stabilization of the negative charge. The other protein effect, is gating mechanism which is created from the conformational changes of Leucine 41. The leucine 41 has a non-polar side chain that allows for transient penetration of the water molecules.[3] This increases the polarity of the redox site environment. The Leucine 41 side chain has two different conformations; reduced and the oxidized form.[4] The conformation in the reduced form is open and allows water molecules near the [Fe(S-Cys)4] 2+ active site and stabilizing the higher net positive charge of the reduced Fe 2+ oxidation state. This shifts the potential 50 mV more positive as indicated by Leucine 41 – Alanine site directed mutagenesis shift of the Fe 3+/2+ redox potential 50 mV more positive.[4] The conformation allows for the infiltration of water molecules which lets the formation of the strongly Hydrogen-bond to attach.[3]
References
- Calderon RH, García-Cerdán JG, Malnoë A, Cook R, Russell JJ, Gaw C, et al. (September 2013). "A conserved rubredoxin is necessary for photosystem II accumulation in diverse oxygenic photoautotrophs". The Journal of Biological Chemistry. 288 (37): 26688–96. doi:10.1074/jbc.M113.487629. PMC 3772215. PMID 23900844.
- Rose K, Shadle SE, Eidsness MK, Kurtz DM, Scott RA, Hedman B, Hodgson KO, Solomon EI (October 1998). "Investigation of Iron−Sulfur Covalency in Rubredoxins and a Model System Using Sulfur K-Edge X-ray Absorption Spectroscopy". Journal of the American Chemical Society. 120 (41): 10743–10747. doi:10.1021/ja981350c. ISSN 0002-7863.
- Min T, Ergenekan CE, Eidsness MK, Ichiye T, Kang C (March 2001). "Leucine 41 is a gate for water entry in the reduction of Clostridium pasteurianum rubredoxin". Protein Science. 10 (3): 613–21. doi:10.1110/gad.34501. PMC 2374124. PMID 11344329.
- Park IY, Youn B, Harley JL, Eidsness MK, Smith E, Ichiye T, Kang C (June 2004). "The unique hydrogen bonded water in the reduced form of Clostridium pasteurianum rubredoxin and its possible role in electron transfer". Journal of Biological Inorganic Chemistry. 9 (4): 423–8. doi:10.1007/s00775-004-0542-3. PMID 15067525.
