Synthesis-dependent strand annealing
Synthesis-dependent strand annealing (SDSA) is a major mechanism of homology-directed repair of DNA double-strand breaks (DSBs). Although many of the features of SDSA were first suggested in 1976[1], the double-Holliday junction model proposed in 1983[2] was favored by many researchers. In 1994, studies of double-strand gap repair in Drosophila were found to be incompatible with the double-Holliday junction model, leading researchers to propose a model they called synthesis-dependent strand annealing.[3] Subsequent studies of meiotic recombination in S. cerevisiae found that non-crossover products appear earlier than double-Holliday junctions or crossover products, challenging the previous notion that both crossover and non-crossover products are produced by double-Holliday junctions and leading the authors to propose that non-crossover products are generated through SDSA.[4]
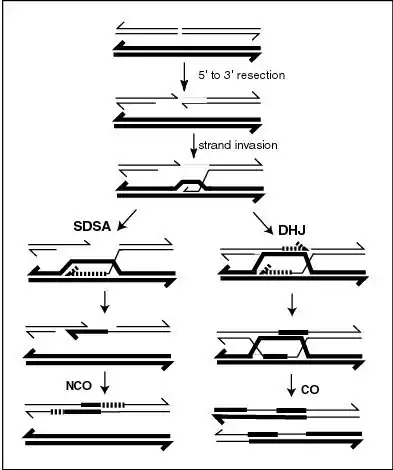
In the SDSA model, repair of double-stranded breaks occurs without the formation of a double Holliday junction, so that the two processes of homologous recombination are identical until just after D-loop formation.[5] In yeast, the D-loop is formed by strand invasion with the help of proteins Rad51 and Rad52,[6] and is then acted on by DNA helicase Srs2 to prevent formation of the double Holliday junction in order for the SDSA pathway to occur.[7] The invading 3' strand is thus extended along the recipient homologous DNA duplex by DNA polymerase in the 5' to 3' direction, so that the D-loop physically translocates – a process referred to as bubble migration DNA synthesis.[8] The resulting single Holliday junction then slides down the DNA duplex in the same direction in a process called branch migration, displacing the extended strand from the template strand. This displaced strand pops up to form a 3' overhang in the original double-stranded break duplex, which can then anneal to the opposite end of the original break through complementary base pairing. Thus DNA synthesis fills in gaps left over from annealing, and extends both ends of the still present single stranded DNA break, ligating all remaining gaps to produce recombinant non-crossover DNA.[9]
SDSA is unique in that D-loop translocation results in conservative rather than semiconservative replication, as the first extended strand is displaced from its template strand, leaving the homologous duplex intact. Therefore, although SDSA produces non-crossover products because flanking markers of heteroduplex DNA are not exchanged, gene conversion may occur, wherein nonreciprocal genetic transfer takes place between two homologous sequences.[10]
Enzymes employed in SDSA during meiosis
Assembly of a nucleoprotein filament comprising single-stranded DNA (ssDNA) and the RecA homolog, Rad51, is a key step necessary for homology search during recombination. In the budding yeast Saccharomyces cerevisiae, Srs2 translocase dismantles Rad51 filaments during meiosis.[11] By directly interacting with Rad51, Srs2 dislodges Rad51 from nucleoprotein filaments thereby inhibiting Rad51-dependent formation of joint molecules and D-loop structures. This dismantling activity is specific for Rad51 since Srs2 does not dismantle DMC1 (a meiosis-specific Rad51 homolog), Rad52 (a Rad 51 mediator) or replication protein A (RPA, a single-stranded DNA binding protein). Srs2 promotes the non-crossover SDSA pathway, apparently by regulating RAD51 binding during strand exchange.[12]
Divergence between SDSA and double-Holliday junction occurs when the D-loop is disassembled allow the nascent strand to anneal to other resected end of the DSB (in the double-Holliday junction model the strand displaced by D-loop extension anneals to the other end of the DSB in "2nd end capture"). Research in Drosophila melanogaster identified the Bloom syndrome helicase (Blm) as the enzyme promoting dissassembly of the D-loop.[13][14][15] Similarly, S. cerevisiae Sgs1, an ortholog of BLM, appears to be a central regulator of most of the recombination events that occur during S. cerevisiae meiosis.[16] Sgs1(BLM) may disassemble D-loop structures analogous to early strand invasion intermediates and thus promote NCO formation by SDSA.[16] The Sgs1 helicase forms a conserved complex with the topoisomerase III (Top3)-RMI1 heterodimer (that catalyzes DNA single strand passage). This complex, called STR (for its three components), promotes early formation of NCO recombinants by SDSA during meiosis.[17]
As reviewed by Uringa et al.[18] the RTEL1 helicase is proposed to regulate recombination during meiosis in the worm Caenorhabditis elegans. RTEL1 is a key protein in repair of DSBs. It disrupts D-loops and is though to promote NCO outcomes through SDSA.
The number of DSBs created during meiosis can substantially exceed the number of final CO events. In the plant Arabidopsis thaliana, only about 4% of DSBs are repaired by CO recombination,[19] suggesting that most DSBs are repaired by NCO recombination. Data based on tetrad analysis from several species of fungi show that only a minority (on average about 34%) of recombination events during meiosis are COs (see Whitehouse,[20] Tables 19 and 38 for summaries of data from S. cerevisiae, Podospora anserina, Sordaria fimicola and Sordaria brevicollis). In the fruit fly D. melanogaster during meiosis in females there is at least a 3:1 ratio of NCOs to COs.[21] These observations indicate that the majority of recombination events during meiosis are NCOs, and suggest that SDSA is the principal pathway for recombination during meiosis.
References
- Resnick MA (June 1976). "The repair of double-strand breaks in DNA; a model involving recombination". Journal of Theoretical Biology. 59 (1): 97–106. doi:10.1016/s0022-5193(76)80025-2. PMID 940351.
- Szostak JW, Orr-Weaver TL, Rothstein R, Stahl FW (May 1983). "The double-strand-break repair model for recombination". Cell. 33 (1): 25–35. doi:10.1016/0092-8674(83)90331-8.
- Nassif N, Penney J, Pal S, Engels WR, Gloor GB (March 1994). "Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair". Molecular and Cellular Biology. 14 (3): 1613–25. doi:10.1128/mcb.14.3.1613. PMC 358520. PMID 8114699.
- Allers T, Lichten M (July 2001). "Differential timing and control of noncrossover and crossover recombination during meiosis". Cell. 106 (1): 47–57. doi:10.1016/s0092-8674(01)00416-0. PMID 11461701.
- McMahill MS, Sham CW, Bishop DK (November 2007). "Synthesis-dependent strand annealing in meiosis". PLoS Biology. 5 (11): e299. doi:10.1371/journal.pbio.0050299. PMC 2062477. PMID 17988174.
- Dupaigne P, Le Breton C, Fabre F, Gangloff S, Le Cam E, Veaute X (February 2008). "The Srs2 helicase activity is stimulated by Rad51 filaments on dsDNA: implications for crossover incidence during mitotic recombination". Molecular Cell. 29 (2): 243–54. doi:10.1016/j.molcel.2007.11.033. PMID 18243118.
- Miura T, Yamana Y, Usui T, Ogawa HI, Yamamoto MT, Kusano K (May 2012). "Homologous recombination via synthesis-dependent strand annealing in yeast requires the Irc20 and Srs2 DNA helicases". Genetics. 191 (1): 65–78. doi:10.1534/genetics.112.139105. PMC 3338270. PMID 22367032.
- Formosa T, Alberts BM (December 1986). "DNA synthesis dependent on genetic recombination: characterization of a reaction catalyzed by purified bacteriophage T4 proteins". Cell. 47 (5): 793–806. doi:10.1016/0092-8674(86)90522-2. PMID 3022939.
- Helleday T, Lo J, van Gent DC, Engelward BP (July 2007). "DNA double-strand break repair: from mechanistic understanding to cancer treatment". DNA Repair. 6 (7): 923–35. doi:10.1016/j.dnarep.2007.02.006. PMID 17363343.
- Maher RL, Branagan AM, Morrical SW (October 2011). "Coordination of DNA replication and recombination activities in the maintenance of genome stability". Journal of Cellular Biochemistry. 112 (10): 2672–82. doi:10.1002/jcb.23211. PMC 3178728. PMID 21647941.
- Sasanuma H, Furihata Y, Shinohara M, Shinohara A (August 2013). "Remodeling of the Rad51 DNA strand-exchange protein by the Srs2 helicase". Genetics. 194 (4): 859–72. doi:10.1534/genetics.113.150615. PMC 3730916. PMID 23770697.
- Ira G, Malkova A, Liberi G, Foiani M, Haber JE (November 2003). "Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast". Cell. 115 (4): 401–11. doi:10.1016/s0092-8674(03)00886-9. PMC 4493758. PMID 14622595.
- Adams MD, McVey M, Sekelsky JJ (January 2003). "Drosophila BLM in double-strand break repair by synthesis-dependent strand annealing". Science. 299 (5604): 265–7. doi:10.1126/science.1077198. PMID 12522255.
- McVey M, Adams M, Staeva-Vieira E, Sekelsky JJ (June 2004). "Evidence for multiple cycles of strand invasion during repair of double-strand gaps in Drosophila". Genetics. 167 (2): 699–705. doi:10.1534/genetics.103.025411. PMC 1470890. PMID 15238522.
- McVey M, Larocque JR, Adams MD, Sekelsky JJ (November 2004). "Formation of deletions during double-strand break repair in Drosophila DmBlm mutants occurs after strand invasion". Proceedings of the National Academy of Sciences of the United States of America. 101 (44): 15694–9. doi:10.1073/pnas.0406157101. PMC 524851. PMID 15501916.
- De Muyt A, Jessop L, Kolar E, Sourirajan A, Chen J, Dayani Y, Lichten M (April 2012). "BLM helicase ortholog Sgs1 is a central regulator of meiotic recombination intermediate metabolism". Molecular Cell. 46 (1): 43–53. doi:10.1016/j.molcel.2012.02.020. PMC 3328772. PMID 22500736.
- Kaur H, De Muyt A, Lichten M (February 2015). "Top3-Rmi1 DNA single-strand decatenase is integral to the formation and resolution of meiotic recombination intermediates". Molecular Cell. 57 (4): 583–594. doi:10.1016/j.molcel.2015.01.020. PMC 4338413. PMID 25699707.
- Uringa EJ, Youds JL, Lisaingo K, Lansdorp PM, Boulton SJ (March 2011). "RTEL1: an essential helicase for telomere maintenance and the regulation of homologous recombination". Nucleic Acids Research. 39 (5): 1647–55. doi:10.1093/nar/gkq1045. PMC 3061057. PMID 21097466.
- Crismani W, Girard C, Froger N, Pradillo M, Santos JL, Chelysheva L, et al. (June 2012). "FANCM limits meiotic crossovers". Science. 336 (6088): 1588–90. Bibcode:2012Sci...336.1588C. doi:10.1126/science.1220381. PMID 22723424.
- Whitehouse, HLK (1982). Genetic Recombination: understanding the mechanisms. Wiley. p. 321 & Table 38. ISBN 978-0471102052.
- Mehrotra S, McKim KS (November 2006). "Temporal analysis of meiotic DNA double-strand break formation and repair in Drosophila females". PLoS Genetics. 2 (11): e200. doi:10.1371/journal.pgen.0020200. PMC 1657055. PMID 17166055.