Carnitine O-acetyltransferase
Carnitine O-acetyltransferase also called carnitine acetyltransferase (CRAT, or CAT)[5] (EC 2.3.1.7) is an enzyme that encoded by the CRAT gene that catalyzes the chemical reaction
- acetyl-CoA + carnitine CoA + acetylcarnitine
where the acetyl group displaces the hydrogen atom in the central hydroxyl group of carnitine.[6]
Thus, the two substrates of this enzyme are acetyl-CoA and carnitine, whereas its two products are CoA and O-acetylcarnitine. The reaction is highly reversible and does not depend on the order in which substrates bind.[6]
Different subcellular localizations of the CRAT mRNAs are thought to result from alternative splicing of the CRAT gene suggested by the divergent sequences in the 5' region of peroxisomal and mitochondrial CRAT cDNAs and the location of an intron where the sequences diverge. The alternatively splicing of this gene results in three distinct isoforms, one of which contains an N-terminal mitochondrial transit peptide, and has been shown to be located in mitochondria.[7]
Nomenclature
This enzyme belongs to the family of transferases, to be specific those acyltransferases transferring groups other than aminoacyl groups. The systematic name of this enzyme class is acetyl-CoA:carnitine O-acetyltransferase. Other names in common use include acetyl-CoA-carnitine O-acetyltransferase, acetylcarnitine transferase, carnitine acetyl coenzyme A transferase, carnitine acetylase, carnitine acetyltransferase, carnitine-acetyl-CoA transferase, and CATC. This enzyme participates in alanine and aspartate metabolism.
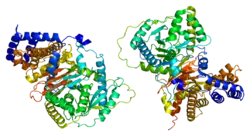
Structure
In general, carnitine acetyltransferases have molecular weights of about 70 kDa, and contain approximately 600 residues1. CRAT contains two domains, an N domain and a C domain, and is composed of 20 α helices and 16 β strands. The N domain consists of an eight-stranded β sheet flanked on both sides by eight α helices. A six-stranded mixed β sheet and eleven α helices comprise the enzyme’s C domain.
When compared, the cores of the two domains reflect significantly similar peptide backbone folding. This occurs despite the fact that only 4% of the amino acids that comprise those peptide backbones corresponds to one another.[5]
Active site
His343 is the catalytic residue in CRAT.[8] It is located at the interface between the enzyme’s C and N domains towards the heart of CRAT. His343 is accessible via two 15-18 Å channels that approach the residue from opposite ends of the CRAT enzyme. These channels are utilized by the substrates of CRAT, one channel for carnitine, and one for CoA. The side chain of His343 is positioned irregularly, with the δ1 ring nitrogen hydrogen bonded to the carbonyl oxygen on the amino acid backbone.[5][9][10]
CoA binding site
Due to the fact that CRAT binds CoA, rather than acetyl-CoA, it appears that CRAT possesses the ability to hydrolyze acetyl-CoA, before interacting with the lone CoA fragment at the binding site.[5] CoA is bound in a linear conformation with its pantothenic arm binding at the active site. Here, the pantothenic arm’s terminal thiol group and the ε2 nitrogen on the catalytic His343 side chain form a hydrogen bond. The 3’-phosphate on CoA forms interactions with residues Lys419 and Lys423. Also at the binding site, the residues Asp430 and Glu453 form a direct hydrogen bond to one another. If either residue exhibits a mutation, can result in a decrease in CRAT activity.[11][12]
Carnitine binding site
Carnitine binds to CRAT in a partially folded state, with its hydroxyl group and carboxyl group facing opposite directions. The site itself is composed of the C domain β sheet and particular residues from the N domain. Upon binding, a face of carnitine is left exposed to the space outside the enzyme. Like CoA, carnitine forms a hydrogen bond with the ε2 nitrogen on His343. In the case of carnitine, the bond is formed with its 3-hydroxyl group. This CRAT catalysis is stereospecific for carnitine, as the stereoisomer of the 3-hydroxyl group cannot sufficiently interact with the CRAT carnitine binding site. CRAT undergoes minor conformational changes upon binding with carnitine.[5][13][14]
Function
Enzyme mechanism
| carnitine O-acetyltransferase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
.tiff.png.webp) transferase Mechanism (His343) | |||||||||
| Identifiers | |||||||||
| EC number | 2.3.1.7 | ||||||||
| CAS number | 9029-90-7 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
The His343 residue at the active site of CRAT acts as a base that is able to deprotonate the CoA thiol group or the Carnitine 3’-hydroxyl group depending on the direction of the reaction. The structure of CRAT optimizes this reaction by causing direct hydrogen bonding between the His343 and both substrates. The deprotonated group is now free to attack the acetyl group of acetyl-CoA or acetylcarnitine at its carbonyl site. The reaction proceeds directly, without the formation of a His343-acetyl intermediate.
Hydrolysis
It is possible for catalysis to occur with only one of the two substrates. If either acetyl-CoA or acetylcarnitine binds to CRAT, a water molecule may fill the other binding site and act as an acetyl group acceptor.
Substrate-assisted catalysis
The literature suggests that the trimethylammonium group on carnitine may be a crucial factor in CRAT catalysis. This group exhibits a positive charge that stabilizes the oxyanion in the reaction’s intermediate. This idea is supported by the fact the positive charge of carnitine is unnecessary for active site binding, but vital for the catalysis to proceed. This has been proven to be the case through the synthesis of a carnitine analog lacking its trimethylammonium group. This compound was able to compete with carnitine in binding to CRAT, but was unable to induce a reaction.[15] The emergence of subtrate-assisted catalysis has opened up new strategies for increasing synthetic substrate specificity.[16]
Biological function
There is evidence that suggests that CRAT activity is necessary for the cell cycle to proceed from the G1 phase to the S phase.[17]
Clinical significance
Those with an inherited deficiency in CRAT activity are at risk for developing severe heart and neurological problems.[5]
Reduced CRAT activity can be found in individuals suffering from Alzheimer’s disease.[5]
CRAT and its family of enzymes have great potential as targets for developing therapeutic treatments for Type 2 diabetes and other diseases.[18][19][20]
References
- GRCh38: Ensembl release 89: ENSG00000095321 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000026853 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Jogl G, Tong L (Jan 2003). "Crystal structure of carnitine acetyltransferase and implications for the catalytic mechanism and fatty acid transport". Cell. 112 (1): 113–22. doi:10.1016/S0092-8674(02)01228-X. PMID 12526798. S2CID 18633987.
- Bieber LL (1988). "Carnitine". Annual Review of Biochemistry. 57: 261–83. doi:10.1146/annurev.bi.57.070188.001401. PMID 3052273.
- "Entrez Gene: CRAT carnitine acetyltransferase".
- McGarry JD, Brown NF (Feb 1997). "The mitochondrial carnitine palmitoyltransferase system. From concept to molecular analysis". European Journal of Biochemistry / FEBS. 244 (1): 1–14. doi:10.1111/j.1432-1033.1997.00001.x. PMID 9063439.
- Jogl G, Hsiao YS, Tong L (Nov 2004). "Structure and function of carnitine acyltransferases". Annals of the New York Academy of Sciences. 1033 (1): 17–29. Bibcode:2004NYASA1033...17J. doi:10.1196/annals.1320.002. PMID 15591000. S2CID 24466239.
- Wu D, Govindasamy L, Lian W, Gu Y, Kukar T, Agbandje-McKenna M, McKenna R (Apr 2003). "Structure of human carnitine acetyltransferase. Molecular basis for fatty acyl transfer". The Journal of Biological Chemistry. 278 (15): 13159–65. doi:10.1074/jbc.M212356200. PMID 12562770.
- Ramsay RR, Gandour RD, van der Leij FR (Mar 2001). "Molecular enzymology of carnitine transfer and transport". Biochimica et Biophysica Acta (BBA) - Protein Structure and Molecular Enzymology. 1546 (1): 21–43. doi:10.1016/S0167-4838(01)00147-9. PMID 11257506.
- Hsiao YS, Jogl G, Tong L (Sep 2006). "Crystal structures of murine carnitine acetyltransferase in ternary complexes with its substrates". The Journal of Biological Chemistry. 281 (38): 28480–7. doi:10.1074/jbc.M602622200. PMC 2940834. PMID 16870616.
- Cronin CN (Sep 1997). "The conserved serine-threonine-serine motif of the carnitine acyltransferases is involved in carnitine binding and transition-state stabilization: a site-directed mutagenesis study". Biochemical and Biophysical Research Communications. 238 (3): 784–9. doi:10.1006/bbrc.1997.7390. PMID 9325168.
- Hsiao YS, Jogl G, Tong L (Jul 2004). "Structural and biochemical studies of the substrate selectivity of carnitine acetyltransferase". The Journal of Biological Chemistry. 279 (30): 31584–9. doi:10.1074/jbc.M403484200. PMID 15155726.
- Saeed A, McMillin JB, Wolkowicz PE, Brouillette WJ (Sep 1993). "Carnitine acyltransferase enzymic catalysis requires a positive charge on the carnitine cofactor". Archives of Biochemistry and Biophysics. 305 (2): 307–12. doi:10.1006/abbi.1993.1427. PMID 8373168.
- Dall'Acqua W, Carter P (Jan 2000). "Substrate-assisted catalysis: molecular basis and biological significance". Protein Science. 9 (1): 1–9. doi:10.1110/ps.9.1.1. PMC 2144443. PMID 10739241.
- Brunner S, Kramar K, Denhardt DT, Hofbauer R (Mar 1997). "Cloning and characterization of murine carnitine acetyltransferase: evidence for a requirement during cell cycle progression". The Biochemical Journal. 322 (2): 403–10. doi:10.1042/bj3220403. PMC 1218205. PMID 9065756.
- Anderson RC (Feb 1998). "Carnitine palmitoyltransferase: a viable target for the treatment of NIDDM?". Current Pharmaceutical Design. 4 (1): 1–16. PMID 10197030.
- Giannessi F, Chiodi P, Marzi M, Minetti P, Pessotto P, De Angelis F, Tassoni E, Conti R, Giorgi F, Mabilia M, Dell'Uomo N, Muck S, Tinti MO, Carminati P, Arduini A (Jul 2001). "Reversible carnitine palmitoyltransferase inhibitors with broad chemical diversity as potential antidiabetic agents". Journal of Medicinal Chemistry. 44 (15): 2383–6. doi:10.1021/jm010889+. PMID 11448219.
- Wagman AS, Nuss JM (Apr 2001). "Current therapies and emerging targets for the treatment of diabetes". Current Pharmaceutical Design. 7 (6): 417–50. doi:10.2174/1381612013397915. PMID 11281851.
Further reading
- Chase JF, Pearson DJ, Tubbs PK (Jan 1965). "The Preparation of Crystallin Carnitine Acetyltransferase". Biochimica et Biophysica Acta (BBA) - Nucleic Acids and Protein Synthesis. 96: 162–5. doi:10.1016/0005-2787(65)90622-2. PMID 14285260.
- Friedman S, Fraenkel G (Dec 1955). "Reversible enzymatic acetylation of carnitine". Archives of Biochemistry and Biophysics. 59 (2): 491–501. doi:10.1016/0003-9861(55)90515-4. PMID 13275966.
- Miyazawa S, Ozasa H, Furuta S, Osumi T, Hashimoto T (Feb 1983). "Purification and properties of carnitine acetyltransferase from rat liver". Journal of Biochemistry. 93 (2): 439–51. doi:10.1093/oxfordjournals.jbchem.a134198. PMID 6404901.
]