Codon degeneracy
Degeneracy of codons is the redundancy of the genetic code, exhibited as the multiplicity of three-base pair codon combinations that specify an amino acid. The degeneracy of the genetic code is what accounts for the existence of synonymous mutations.[1]:Chp 15
Background
Degeneracy of the genetic code was identified by Lagerkvist.[2] For instance, codons GAA and GAG both specify glutamic acid and exhibit redundancy; but, neither specifies any other amino acid and thus are not ambiguous or demonstrate no ambiguity.
The codons encoding one amino acid may differ in any of their three positions; however, more often than not, this difference is in the second or third position.[3] For instance, the amino acid glutamic acid is specified by GAA and GAG codons (difference in the third position); the amino acid leucine is specified by UUA, UUG, CUU, CUC, CUA, CUG codons (difference in the first or third position); and the amino acid serine is specified by UCA, UCG, UCC, UCU, AGU, AGC (difference in the first, second, or third position).[1]:521–522
Degeneracy results because there are more codons than encodable amino acids. For example, if there were two bases per codon, then only 16 amino acids could be coded for (4²=16). Because at least 21 codes are required (20 amino acids plus stop) and the next largest number of bases is three, then 4³ gives 64 possible codons, meaning that some degeneracy must exist.[1]:521–522 The appearance of codon degeneracy implies the existence of certain symmetry for codon multiplicity assignment.[4]
Implications
These properties of the genetic code make it more fault-tolerant for point mutations. For example, in theory, fourfold degenerate codons can tolerate any point mutation at the third position, although codon usage bias restricts this in practice in many organisms; twofold degenerate codons can withstand silence mutation rather than Missense or Nonsense point mutations at the third position. Since transition mutations (purine to purine or pyrimidine to pyrimidine mutations) are more likely than transversion (purine to pyrimidine or vice versa) mutations, the equivalence of purines or that of pyrimidines at twofold degenerate sites adds a further fault-tolerance.[1]:531–532
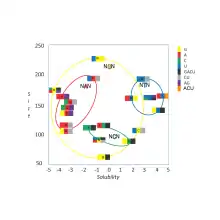
A practical consequence of redundancy is that some errors in the genetic code cause only a silent mutation or an error that would not affect the protein because the hydrophilicity or hydrophobicity is maintained by equivalent substitution of amino acids; for example, a codon of NUN (where N = any nucleotide) tends to code for hydrophobic amino acids. NCN yields amino acid residues that are small in size and moderate in hydropathy; NAN encodes average size hydrophilic residues.[5][6] These tendencies may result from the shared ancestry of the aminoacyl tRNA synthetases related to these codons.
These variable codes for amino acids are allowed because of modified bases in the first base of the anticodon of the tRNA, and the base-pair formed is called a wobble base pair. The modified bases include inosine and the Non-Watson-Crick U-G basepair.[7]
Terminology
A position of a codon is said to be a n-fold degenerate site if only n of four possible nucleotides (A, C, G, T) at this position specify the same amino acid. A nucleotide substitution at a fourfold degenerate site is referred to as a synonymous nucleotide substitution,[1]:521–522 whereas nucleotide substitutions in which the substitution involves the change of a purine to a pyrimidine, or vice versa, are non-synonymous transversion substitutions.[1]:521–522
A position of a codon is said to be a non-degenerate site if any mutation at this position results in amino acid substitution. There is only one threefold degenerate site where changing to three of the four nucleotides may have no effect on the amino acid (depending on what it is changed to), while changing to the fourth possible nucleotide always results in an amino acid substitution. This is the third position of an isoleucine codon: AUU, AUC, or AUA all encode isoleucine, but AUG encodes methionine. In computation, this position is often treated as a twofold degenerate site.[1]:521–522
There are three amino acids encoded by six different codons: serine, leucine, and arginine. Only two amino acids are specified by a single codon each. One of these is the amino-acid methionine, specified by the codon AUG, which also specifies the start of translation; the other is tryptophan, specified by the codon UGG.
| Amino acid | DNA codons | Compressed | Amino acid | DNA codons | Compressed | |
|---|---|---|---|---|---|---|
| Ala, A | GCU, GCC, GCA, GCG | GCN | Ile, I | AUU, AUC, AUA | AUH | |
| Arg, R | CGU, CGC, CGA, CGG; AGA, AGG | CGN, AGR; or CGY, MGR |
Leu, L | CUU, CUC, CUA, CUG; UUA, UUG | CUN, UUR; or CUY, YUR | |
| Asn, N | AAU, AAC | AAY | Lys, K | AAA, AAG | AAR | |
| Asp, D | GAU, GAC | GAY | Met, M | AUG | ||
| Asn or Asp, B | AAU, AAC; GAU, GAC | RAY | Phe, F | UUU, UUC | UUY | |
| Cys, C | UGU, UGC | UGY | Pro, P | CCU, CCC, CCA, CCG | CCN | |
| Gln, Q | CAA, CAG | CAR | Ser, S | UCU, UCC, UCA, UCG; AGU, AGC | UCN, AGY | |
| Glu, E | GAA, GAG | GAR | Thr, T | ACU, ACC, ACA, ACG | ACN | |
| Gln or Glu, Z | CAA, CAG; GAA, GAG | SAR | Trp, W | UGG | ||
| Gly, G | GGU, GGC, GGA, GGG | GGN | Tyr, Y | UAU, UAC | UAY | |
| His, H | CAU, CAC | CAY | Val, V | GUU, GUC, GUA, GUG | GUN | |
| START | AUG | STOP | UAA, UGA, UAG | URA, UAR | ||
References
- Watson JD, Baker TA, Bell SP, Gann A, Levine M, Oosick R (2008). Molecular Biology of the Gene. San Francisco: Pearson/Benjamin Cummings. ISBN 978-0-8053-9592-1.
- Lagerkvist, U. (1978.) "Two out of three: An alternative method for codon reading", PNAS, 75:1759-62.
- Lehmann, J; Libchaber, A (July 2008). "Degeneracy of the genetic code and stability of the base pair at the second position of the anticodon". RNA. 14 (7): 1264–9. doi:10.1261/rna.1029808. PMC 2441979. PMID 18495942.
- Shu, Jian-Jun (2017). "A new integrated symmetrical table for genetic codes". BioSystems. 151: 21–26. arXiv:1703.03787. doi:10.1016/j.biosystems.2016.11.004. PMID 27887904.
- Yang; et al. (1990). Michel-Beyerle, M. E. (ed.). Reaction centers of photosynthetic bacteria: Feldafing-II-Meeting. 6. Berlin: Springer-Verlag. pp. 209–18. ISBN 3-540-53420-2.
- Füllen G, Youvan DC (1994). "Genetic Algorithms and Recursive Ensemble Mutagenesis in Protein Engineering". Complexity International. 1. Archived from the original on 2011-03-15.
- Varani G, McClain WH (July 2000). "The G x U wobble base pair. A fundamental building block of RNA structure crucial to RNA function in diverse biological systems". EMBO Rep. 1 (1): 18–23. doi:10.1093/embo-reports/kvd001. PMC 1083677. PMID 11256617.
