Guide RNA
Guide RNAs (a.k.a. gRNA, sgRNA) are the RNAs that guide the insertion or deletion of uridine residues into mitochondrial mRNAs in kinetoplastid protists in a process known as RNA editing.[1]
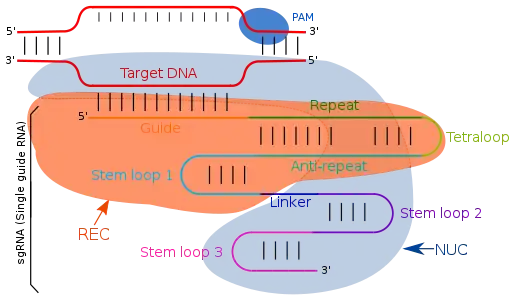
The terms "guide RNA" and "gRNA" are also used in prokaryotic DNA editing involving CRISPR and Cas9. For this prokaryotic DNA-editing system, the gRNA confers target sequence specificity to the CRISPR-Cas9 system. These gRNAs are non coding short RNA sequences which bind to the complementary target DNA sequences. Guide RNA first binds to the Cas9 enzyme and the gRNA sequence guides the complex via pairing to a specific location on the DNA, where Cas9 performs its endonuclease activity by cutting the target DNA strand.
In addition to expression of the Cas9 nuclease, the CRISPR-Cas9 system requires a specific RNA molecule to recruit and direct the nuclease activity to the region of interest. These guide RNAs take one of two forms:
- A synthetic trans-activating CRISPR RNA (tracrRNA) plus a synthetic CRISPR RNA (crRNA) designed to cleave the gene target site of interest
- A synthetic or expressed single guide RNA (sgRNA) that consists of both the crRNA and tracrRNA as a single construct
The crRNA and the tracrRNA form a complex which acts as the guide RNA for the Cas9 enzyme. The scaffolding ability of tracrRNA along with crRNA specificity can be combined into a single synthetic gRNA which simplifies guiding of gene alterations to a one component system which may increase efficiencies.
History
The RNA-editing Guide RNA was discovered in 1990 by B. Blum, N. Bakalara, and L. Simpson[2] because of their role in RNA editing in the mitochondrion of Leishmania tarentolae. These gRNA molecules are encoded in maxicircle DNA in mitochondria having sequences that are complementary to mature mRNAs within the edited regions. They participate in multiple activities to cleave, insert, or delete bases after the formation of partial hybrid between gRNA and pre-edited mRNA
Guide RNA in Protists
Trypanosomatid protists and other kinetoplastids have a novel post-transcriptional mitochondrial RNA modification process known as "RNA editing". They have large segment of highly organized DNA segments present in mitochondria. This mitochondrial DNA is circular and exists in one of two forms, maxicircles or minicircles. There are 20-50 maxicircles per cells having both coding and non coding regions. The coding region is highly conserved (16-17kb) and non coding region varies depending on the species. Minicircles are small but more numerous than maxicircles. Minicircles constitute 95% of the mass of kinetoplastid DNA. Maxicircles can encode "cryptogenes" and some gRNAs; minicircles can encode the majority of gRNAs. As many as 1000 gRNAs can be encoded by 250 or more minicircles. Some gRNA genes show identical insertion and deletion sites even if they have different sequences, whereas other gRNA sequences are not complementary to pre-edited mRNA. Maxicircles and minicircles molecules are catenated into a giant network of DNA that is situated at the base of the flagellum in the inner compartment of the single mitochondrion.[2]
A majority of the maxicircle transcripts can not be translated into proteins due to multiple frameshifts in the sequences. These frameshifts are corrected after transcription by the insertion and deletion of uridine residues at precise sites which create an open reading frame that is translated into a mitochondrial protein homologous to mitochondrial proteins from other cells. The insertions and deletions are mediated by short guide RNA (gRNAs) which encode the editing information in the form of complementary sequences (allowing GU as well as GC base pairs).
gRNA-mRNA Complex
The guide RNA are mainly transcribed from the intergenic region of DNA maxicircle and these are complementary to mature mRNA. It is important for gRNA to interact initially with pre-edited mRNA and then its 5' region base pair with complementary mRNA . The 3' end of gRNA contains oligo 'U' tail (5-25 nucleotides in length) which is a non encoded region but interacts and forms a stable complex with A and G rich region of mRNA. This initial hybrid helps in the recognition of specific mRNA site to be edited.[3]
Function
The presence of two genomes in the mitochondrion, one of which contains sequence information that corrects errors in the other genome, is novel. Editing proceeds generally 3' to 5' on the mRNA. The initial editing event occurs when a gRNA forms an RNA duplex with a complementary mRNA sequence just downstream of the editing site. This then recruits a number of ribonucleoprotein complexes that direct the cleavage of first mismatched base adjacent to gRNA-mRNA anchor. Uridyly transferase inserts 'U' at 3' terminal and RNA ligase is responsible for joining of two cut ends. The adjacent upstream editing site is then modified in the same manner. A single gRNA usually encodes the information for several editing sites (an editing "block"), the editing of which produces a complete gRNA/mRNA duplex. This process of modification is termed as original enzyme cascade model.[4]
In the case of "pan-edited" mRNAs,[5] the duplex unwinds and another gRNA then forms a duplex with the edited mRNA sequence and initiates another round of editing. The overlapping gRNAs form an editing "domain". In some genes there are multiple editing domains. The extent of editing for any particular gene varies between trypanosomatid species. The variation consists of the loss of editing at the 3' side, probably due to the loss of minicircle sequence classes that encode specific gRNAs. A retroposition model has been proposed to account for the partial, and in some cases, complete, loss of editing in evolution. Loss of editing is lethal in most cases, although losses have been seen in old laboratory strains. The maintenance of editing over the long evolutionary history of these ancient protists suggests the presence of a selective advantage, the exact nature of which is still uncertain.
It is not clear why trypanosomatids utilize such an elaborate mechanism to produce mRNAs. It may have originated in the early mitochondrion of the ancestor of the kintoplastid protist lineage, since it is present in the bodonids which are ancestral to the trypanosomatids, and may not be present in the euglenoids, which branched from the same common ancestor as the kinetoplastids.
In the protozoan Leishmania tarentolae, 12 of the 18 mitochondrial genes are edited using this process. One such gene is Cyb. The mRNA is actually edited twice in succession. For the first edit, the relevant sequence on the mRNA is as follows:
mRNA 5' AAAGAAAAGGCUUUAACUUCAGGUUGU 3'
The 3' end is used to anchor the gRNA (gCyb-I gRNA in this case) by basepairing (some G/U pairs are used). The 5' end does not exactly match and one of three specific endonucleases cleaves the mRNA at the mismatch site.
gRNA 3' AAUAAUAAAUUUUUAAAUAUAAUAGAAAAUUGAAGUUCAGUA 5' mRNA 5' A A AGAAA A G G C UUUAACUUCAGGUUGU 3'
The mRNA is now "repaired" by adding U's at each editing site in succession, giving the following sequence:
gRNA 3' AAUAAUAAAUUUUUAAAUAUAAUAGAAAAUUGAAGUUCAGUA 5' mRNA 5' UUAUUAUUUAGAAAUUUAUGUUGUCUUUUAACUUCAGGUUGU 3'
This particular gene has two overlapping gRNA editing sites. The 5' end of this section is the 3' anchor for another gRNA (gCyb-II gRNA)
Guide RNA in Prokaryotes
CRISPR In Prokaryotes
Most of the prokaryotes like bacteria and archea makes the use of their adaptive immune system using CRISPR (clustered regularly interspaced short palindromic repeats) and cas enzyme to detect and remove the foreign genetic material. When prokaryotes are infected by bacteriophages, then the phage DNA give rise to short cluster repeats (CRISPR) which are used to detect and cleave the DNA fragments from similar type of phages. This defense mechanism of prokaryotes is used as editing technique which can used in gene therapy process as well. The CRISPR Cas editing method makes the use gRNA for identification and cleavage of DNA strands.
Structure
Guide RNA targets the complementary sequences by simple Watson-Crick base pairing. In type II CRISPR/cas system, single guide RNA directs the target specific regions. Single guide RNA are artificially programmed combination of two RNA molecules, one component (tracrRNA) is responsible for Cas9 endonuclease activity and other (crRNA) binds to the target specific DNA region. Therefore, the trans activating RNA (tracrRNA) and crRNA are two key components and are joined by tetraloop which results in formation of sgRNA. TracrRNA are base pairs having a stemloop structure in itself and attaches to the endonuclease enzyme. Transcription of CRISPR locus gives CRISPR RNA (crRNA) which have spacer flanked region due to repeat sequences, consisting of 18-20 base pair. crRNA identifies the specific complementary target region which is cleaved by Cas9 after its binding with crRNA and tcRNA, which all together known as effector complex. With the modifications in the crRNA sequences of the guide RNA, the binding location can be changed and hence defining it as a user defined program.
Applications
Designing gRNAs
The targeting specificity of CRISPR-Cas9 is determined by the 20-nt sequence at the 5' end of the gRNA. The desired target sequence must precede the protospacer adjacent motif (PAM) which is a short DNA sequence usually 2-6 base pairs in length that follows the DNA region targeted for cleavage by the CRISPR system, such as CRISPR-Cas9. The PAM is required for a Cas nuclease to cut and is generally found 3-4 nucleotides downstream from the cut site. After base pairing of the gRNA to the target, Cas9 mediates a double strand break about 3-nt upstream of PAM.
The GC content of the guide sequence should be 40-80%. High GC content stabilizes the RNA-DNA duplex while destabilizing off-target hybridization. The length of the guide sequence should be between 17-24bp noting a shorter sequence minimizes off-target effects. Guide sequences less than 17bp have a chance of targeting multiple loci.
CRISPR Cas9
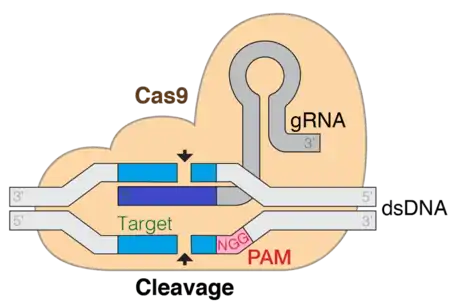
CRISPR (Clustered regularly interspaced short palindromic repeats)/Cas9 is a technique used for gene editing and gene therapy. Cas is an endonuclease enzyme that cuts the DNA at a specific location directed by a guide RNA. This is a target-specific technique that can introduce gene knock out or knock in depending on the double strand repair pathway. Evidence shows that both in-vitro and in-vivo required tracrRNA for Cas9 and target DNA sequence binding. Three main stages constitute CRISPR CAS9 system. The first stage is extension of bases in the CRISPR locus region by addition of foreign DNA spacers in the genome sequence. Several different proteins, like cas1 and cas2, help in finding new spacers. The next stage involves transcription of CRISPR: pre-crRNA (precursor CRISPR RNA) are expressed by the transcription of CRISPR repeat-spacer array. On further modification in the pre-crRNA are converted to single spacer flanked region forming short crRNA. RNA maturation process is similar in type I and II but different in type III, aRNA as tracer are added in this step. The third stage involves binding of cas9 protein and directing it to cleave the DNA segment. Cas9 protein binds to combined form of crRNA and tracrRNA forming an effector complex. This act as guide RNA for cas9 protein directing it for its endonuclease activity.[6]
RNA mutagenesis
One important gene regulation method is RNA mutagenesis which can be introduced by RNA editing with the help of gRNA. Guide RNA replaces adenosine with inosine at the specific target site and modify the genetic code.[7] Adenosine deaminase acts on RNA bringing post transcriptional modification by altering the codons and different protein functions. Guide RNAs are the small nucleolar RNA, these along with riboproteins perform intracellular RNA alterations such as ribomethylation in rRNA and introduction of pseudouridine in preribosomal RNA. Guide RNAs binds to the anti sense RNA sequence and regulates the RNA modification. It is observed that small interfering RNA (siRNA) and micro RNA (miRNA) are generally used as target RNA sequence and modifications are comparatively easy to introduce because of small size.
References
- Hajduk, S. L.; Harris, M. E.; Pollard, V. W. (January 1993). "RNA editing in kinetoplastid mitochondria". FASEB Journal. 7 (1): 54–63. doi:10.1096/fasebj.7.1.8422975. ISSN 0892-6638. PMID 8422975.
- Blum, B.; Bakalara, N.; Simpson, L. (1990-01-26). "A model for RNA editing in kinetoplastid mitochondria: "guide" RNA molecules transcribed from maxicircle DNA provide the edited information". Cell. 60 (2): 189–198. doi:10.1016/0092-8674(90)90735-w. ISSN 0092-8674. PMID 1688737.
- Connell, Gregory J.; Byrne, Elaine M.; Simpson, Larry (1997-02-14). "Guide RNA-independent and Guide RNA-dependent Uridine Insertion into Cytochrome b mRNA in a Mitochondrial Lysate from Leishmania tarentolae ROLE OF RNA SECONDARY STRUCTURE". Journal of Biological Chemistry. 272 (7): 4212–4218. doi:10.1074/jbc.272.7.4212. ISSN 0021-9258. PMID 9020135.
- Connell, Gregory J.; Byrne, Elaine M.; Simpson, Larry (1997-02-14). "Guide RNA-independent and Guide RNA-dependent Uridine Insertion into Cytochrome b mRNA in a Mitochondrial Lysate from Leishmania tarentolae ROLE OF RNA SECONDARY STRUCTURE". Journal of Biological Chemistry. 272 (7): 4212–4218. doi:10.1074/jbc.272.7.4212. ISSN 0021-9258. PMID 9020135.
- Maslov, Dmitri A. (October 2010). "Complete set of mitochondrial pan-edited mRNAs in Leishmania mexicana amazonensis LV78". Molecular and Biochemical Parasitology. 173 (2): 107–114. doi:10.1016/j.molbiopara.2010.05.013. ISSN 0166-6851. PMC 2913609. PMID 20546801.
- Karvelis, Tautvydas; Gasiunas, Giedrius; Miksys, Algirdas; Barrangou, Rodolphe; Horvath, Philippe; Siksnys, Virginijus (2013-05-01). "crRNA and tracrRNA guide Cas9-mediated DNA interference in Streptococcus thermophilus". RNA Biology. 10 (5): 841–851. doi:10.4161/rna.24203. ISSN 1547-6286. PMC 3737341. PMID 23535272.
- Fukuda, Masatora; Umeno, Hiromitsu; Nose, Kanako; Nishitarumizu, Azusa; Noguchi, Ryoma; Nakagawa, Hiroyuki (2017-02-02). "Construction of a guide-RNA for site-directed RNA mutagenesis utilising intracellular A-to-I RNA editing". Scientific Reports. 7: 41478. Bibcode:2017NatSR...741478F. doi:10.1038/srep41478. ISSN 2045-2322. PMC 5288656. PMID 28148949.
Further reading
- Guide RNA-directed uridine insertion RNA editing in vitrohttp://www.jbc.org/content/272/7/4212.full
- Blum, Beat; Simpson, Larry (1990). "Guide RNAs in kinetoplastid mitochondria have a nonencoded 3′ oligo(U) tail involved in recognition of the preedited region". Cell. 62 (2): 391–397. doi:10.1016/0092-8674(90)90375-O. PMID 1695552.
- Kurata, Morito; Wolf, Natalie K.; Lahr, Walker S.; Weg, Madison T.; Kluesner, Mitchell G.; Lee, Samantha; Hui, Kai; Shiraiwa, Masano; Webber, Beau R.; Moriarity, Branden S. (2018). "Highly multiplexed genome engineering using CRISPR/Cas9 gRNA arrays". PLOS ONE. 13 (9): e0198714. Bibcode:2018PLoSO..1398714K. doi:10.1371/journal.pone.0198714. PMC 6141065. PMID 30222773.
- Khan, Fehad J.; Yuen, Garmen; Luo, Ji (2019). "Multiplexed CRISPR/Cas9 gene knockout with simple crRNA:tracrRNA co-transfection". Cell & Bioscience. 9: 41. doi:10.1186/s13578-019-0304-0. PMC 6528186. PMID 31139343.
- Nishimasu, Hiroshi; Nureki, Osamu (2017). "Structures and mechanisms of CRISPR RNA-guided effector nucleases". Current Opinion in Structural Biology. 43: 68–78. doi:10.1016/j.sbi.2016.11.013. PMID 27912110.
- Chuai, Guohui; Ma, Hanhui; Yan, Jifang; Chen, Ming; Hong, Nanfang; Xue, Dongyu; Zhou, Chi; Zhu, Chenyu; Chen, Ke; Duan, Bin; Gu, Feng; Qu, Sheng; Huang, Deshuang; Wei, Jia; Liu, Qi (2018). "DeepCRISPR: Optimized CRISPR guide RNA design by deep learning". Genome Biology. 19 (1): 80. doi:10.1186/s13059-018-1459-4. PMC 6020378. PMID 29945655.
