HCS clustering algorithm
The HCS (Highly Connected Subgraphs) clustering algorithm[1] (also known as the HCS algorithm, and other names such as Highly Connected Clusters/Components/Kernels) is an algorithm based on graph connectivity for cluster analysis. It works by representing the similarity data in a similarity graph, and then finding all the highly connected subgraphs. It does not make any prior assumptions on the number of the clusters. This algorithm was published by Erez Hartuv and Ron Shamir in 2000.
| Class | Cluster analysis (on a similarity graph) |
|---|---|
| Data structure | Graph |
| Worst-case performance | O(2N x f(n,m)) |
| Worst-case space complexity | {{{space}}} |
The HCS algorithm gives a clustering solution, which is inherently meaningful in the application domain, since each solution cluster must have diameter 2 while a union of two solution clusters will have diameter 3.
Similarity modeling and preprocessing
The goal of cluster analysis is to group elements into disjoint subsets, or clusters, based on similarity between elements, so that elements in the same cluster are highly similar to each other (homogeneity), while elements from different clusters have low similarity to each other (separation). Similarity graph is one of the models to represent the similarity between elements, and in turn facilitate generating of clusters. To construct a similarity graph from similarity data, represent elements as vertices, and elicit edges between vertices when the similarity value between them is above some threshold.
Algorithm
In the similarity graph, the more edges exist for a given number of vertices, the more similar such a set of vertices are between each other. In other words, if we try to disconnect a similarity graph by removing edges, the more edges we need to remove before the graph becomes disconnected, the more similar the vertices in this graph. Minimum cut is a minimum set of edges without which the graph will become disconnected.
HCS clustering algorithm finds all the subgraphs with n vertices such that the minimum cut of those subgraphs contain more than n/2 edges, and identifies them as clusters. Such a subgraph is called a Highly Connected Subgraph (HCS). Single vertices are not considered clusters and are grouped into a singletons set S.
Given a similarity graph G(V,E), HCS clustering algorithm will check if it is already highly connected, if yes, returns G, otherwise uses the minimum cut of G to partition G into two subgraphs H and H', and recursively run HCS clustering algorithm on H and H'.
Example
The following animation shows how the HCS clustering algorithm partitions a similarity graph into three clusters.
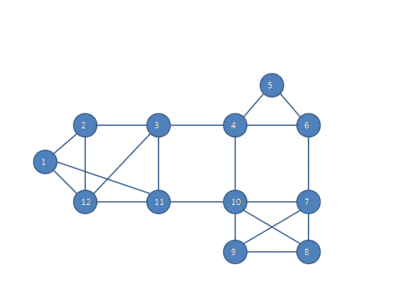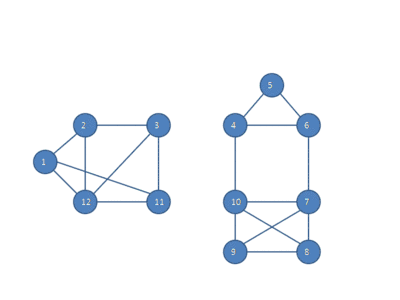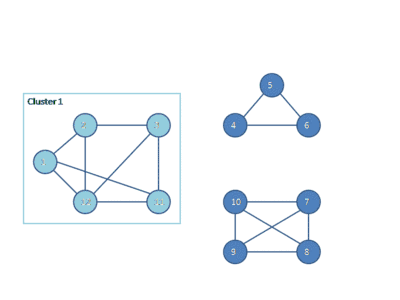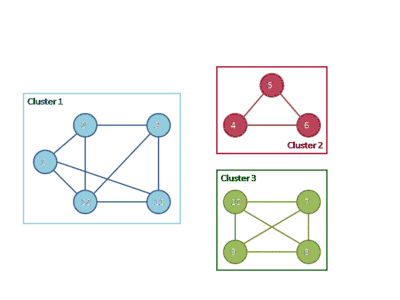
Pseudocode
function HCS(G(V, E)) is
if G is highly connected then
return (G)
else
(H1, H2, C) ← MINIMUMCUT(G)
HCS(H1)
HCS(H2)
end if
end function
The step of finding the minimum cut on graph G is a subroutine that can be implemented using different algorithms for this problem. See below for an example algorithm for finding minimum cut using randomization.
Complexity
The running time of the HCS clustering algorithm is bounded by N × f(n, m). f(n, m) is the time complexity of computing a minimum cut in a graph with n vertices and m edges, and N is the number of clusters found. In many applications N << n.
For fast algorithms for finding a minimum cut in an unweighted graph:
Proofs of properties
The clusters produced by the HCS clustering algorithm possess several properties, which can demonstrate the homogeneity and separation of the solution.
Theorem 1 The diameter of every highly connected graph is at most two.
Proof: Let n=|G|. If G has a vertex x with degree <= n/2, then G has a minimum cut (that isolates x) with edges <= n/2, so G is not highly connected. So if G is highly connected, every vertex has degree >= n/2. There is a famous theorem in graph theory that says that if every vertex has degree >= n/2, then the diameter of G (the longest path between any two nodes) <= 2.
Theorem 2 (a) The number of edges in a highly connected graph is quadratic. (b) The number of edges removed by each iteration of the HCS algorithm is at most linear.
Proof: (a) From Theorem 1 we know that every vertex has degree >= n/2. Therefore, the number of edges in a highly connected graph must be at least (n × n/2)/2, where we sum the degrees of each vertex and divide by 2.
(b) By definition, each iteration removes a minimum cut with <= n/2 edges.
Theorems 1 and 2a provide a strong indication of a final cluster's homogeneity. Doing better approaches the case where all vertices of a cluster are connected, which is both too stringent and also NP-hard.
Theorem 2b indicates separation since any two final clusters C1 and C2 would not have been separated unless there were at most O(C1+C2) edges between them (contrast with the quadratic edges within clusters).
Variations
Singletons adoption: Elements left as singletons by the initial clustering process can be "adopted" by clusters based on similarity to the cluster. If the maximum number of neighbors to a specific cluster is large enough, then it can be added to that cluster.
Removing Low Degree Vertices: When the input graph has vertices with low degrees, it is not worthy to run the algorithm since it is computationally expensive and not informative. Alternatively, a refinement of the algorithm can first remove all vertices with a degree lower than certain threshold.
Examples of HCS usage
- Gene expression analysis[2] The hybridization of synthetic oligonucleotides to arrayed cDNAs yields a fingerprint for each cDNA clone. Run HCS algorithm on these fingerprints can identify clones corresponding to the same gene.
- PPI network structure discovery[3] Using HCS clustering to detect dense subnetworks in PPI that may have biological meaning and represent biological processes.
- "Survey of clustering algorithms." Neural Networks, IEEE Transactions [4]
- The CLICK clustering algorithm[5] is an adaptation of HCS algorithm on weighted similarity graphs, where the weight is assigned with a probability flavor.
- https://www.researchgate.net/publication/259350461_Partitioning_Biological_Networks_into_Highly_Connected_Clusters_with_Maximum_Edge_Coverage Partitioning Biological Networks into Highly Connected Clusters with Maximum Edge Coverage][6]
References
- Hartuv, E.; Shamir, R. (2000), "A clustering algorithm based on graph connectivity", Information Processing Letters, 76 (4–6): 175–181, doi:10.1016/S0020-0190(00)00142-3
- E Hartuv, A O Schmitt, J Lange, S Meier-Ewert, H Lehrach, R Shamir. "An algorithm for clustering cDNA fingerprints." Genomics 66, no. 3 (2000): 249-256.
- Jurisica, Igor, and Dennis Wigle. Knowledge Discovery in Proteomics. Vol. 8. CRC press, 2006.
- Xu, Rui, and Donald Wunsch. "Survey of clustering algorithms." Neural Networks, IEEE Transactions on 16, no. 3 (2005): 645-678.
- Sharan, R.; Shamir, R. (2000), "CLICK: A Clustering Algorithm with Applications to Gene Expression Analysis", Proceedings ISMB '00, 8: 307–316C
- Huffner, F.; Komusiewicz, C.; Liebtrau, A; Niedermeier, R (2014), "Partitioning Biological Networks into Highly Connected Clusters with Maximum Edge Coverage", IEEE/ACM Transactions on Computational Biology and Bioinformatics, 11 (3): 455–467, CiteSeerX 10.1.1.377.1900, doi:10.1109/TCBB.2013.177, PMID 26356014
