Hes3 signaling axis
The STAT3-Ser/Hes3 signaling axis is a specific type of intracellular signaling pathway that regulates several fundamental properties of cells.
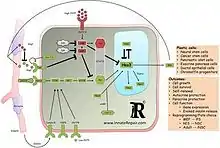
Overview
Cells in tissues need to be able to sense and interpret changes in their environment. For example, cells must be able to detect when they are in physical contact with other cells in order to regulate their growth and avoid the generation of tumors (“carcinogenesis”). In order to do so, cells place receptor molecules on their surface, often with a section of the receptor exposed to the outside of the cell (extracellular environment), and a section inside the cell (intracellular environment). These molecules are exposed to the environment outside of the cell and, therefore, in position to sense it. They are called receptors because when these come into contact with particular molecules (termed ligands), then chemical changes are induced to the receptor. These changes typically involve alterations in the three-dimensional shape of the receptor. These 3D structure changes affect both the extracellular and intracellular parts (domains) of the receptor. As a result, interaction of a receptor with its specific ligand which is located outside of the cell causes changes to the receptor part which is inside the cell. A signal from the extracellular space, therefore, can affect the biochemical state inside the cell.
Following receptor activation by the ligand, several steps can sequentially ensue. For example, the 3D shape changes to the intracellular domain may render it recognizable to catalytic proteins (enzymes) that are located inside the cell and have physical access to it. These enzymes may then induce chemical changes to the intracellular domain of the activated receptor, including the addition of phosphate chemical groups to specific components of the receptor (phosphorylation), or the physical separation (cleavage) of the intracellular domain. Such modifications may enable the intracellular domain to act as an enzyme itself, meaning that it may now catalyze the modification of other proteins in the cell. Enzymes which catalyze phosphorylation modifications are termed kinases. These modified proteins may then also be activated and enabled to induce further modifications to other proteins, and so on. This sequence of catalytic modifications is termed a “signal transduction pathway” or “second messenger cascade”. It is a critical mechanism employed by cells to sense their environment and induce complex changes to their state. Such changes may include, as noted, chemical modifications to other molecules, as well as decisions concerning which genes are activated and which are not (transcriptional regulation).
There are many signal transduction pathways in a cell and each of these involves many different proteins. This provides many opportunities for different signal transduction pathways to intercept (cross-talk). As a result, a cell simultaneously processes and interprets many different signals, as would be expected since the extracellular environment contains many different ligands. Cross-talk also allows the cell to integrate these many signals as opposed to process them independently. For example, mutually opposing signals may be activated at the same time by different ligands, and the cell can interpret these signals as a whole. Signal transduction pathways are widely studied in biology as they provide mechanistic understanding of how a cell operates and takes critical decisions (e.g. to multiply, move, die, activate genes etc.). These pathways also provide many drug targets and are of great relevance to drug discovery efforts.
Technical overview
The notch/STAT3-Ser/Hes3 signaling axis is a recently identified signal transduction branch of the notch[1] signaling pathway, originally shown to regulate the number of neural stem cells in culture and in the living adult brain.[2][3] Pharmacological activation of this pathway opposed the progression of neurodegenerative disease in rodent models. More recent efforts have implicated it in carcinogenesis and diabetes. The pathway can be activated by soluble ligands of the notch receptor which induce the sequential activation of intracellular kinases and the subsequent phosphorylation of STAT3 on the serine residue at amino acid position 727 (STAT3-Ser). This modification is followed by an increase in the levels of Hes3, a transcription factor belonging to the Hes/Hey family of genes (see HES1).[4] Hes3 has been used as a biomarker to identify putative endogenous stem cells in tissues.[5] The pathway is an example of non-canonical signaling as it represents a new branch of a previously established signaling pathway (notch). Several efforts are currently aimed at relating this pathway to other signaling pathways and to manipulate it in a therapeutic context.
Discovery
In canonical notch signaling, ligand proteins bind to the extracellular domain of the notch receptor and induce the cleavage and release of the intracellular domain into the cytoplasm. This subsequently interacts with other proteins, enters the nucleus, and regulates gene expression.[1]
In 2006, a non-canonical branch of the notch signaling pathway was discovered.[2] Using cultures of mouse neural stem cells, notch activation was shown to lead to the phosphorylation of several kinases (PI3K, Akt, mTOR) and subsequent phosphorylation of the serine residue of STAT3 in the absence of any detectable phosphorylation of the tyrosine residue of STAT3, a modification that is widely studied in the context of cancer biology.[6] Following this event, Hes3 mRNA was elevated within 30 minutes. Subsequently, the consequences of this pathway were studied.
Activators
Various inputs into this pathway have been identified. Activators include ligands of a number of receptors. Because certain signal transduction pathways oppose the STAT3-Ser/Hes3 signaling axis, blockers (inhibitors) of these signal transduction pathways promote the STAT3-Ser/Hes3 signaling axis and, therefore, also act as activators:
- A non-canonical branch of the notch signaling pathway (activated by soluble forms of the notch ligands Delta4 and Jagged1). This has been shown in vitro and in vivo.[2]
- Activation of the Tie2 receptor by the ligand Angiopoietin 2. This has been shown in vitro and in vivo.[3][5]
- Activation of the insulin receptor by insulin. This has been shown in vitro and in vivo.[7]
- Treatment with an inhibitor of the Janus kinase (JAK). This has been shown in vitro.[2]
- Treatment with an inhibitor of the p38 MAP kinase kinase. This has been shown in vitro.[2]
- Treatment with cholera toxin. This has been shown in vitro.[8] This particular treatment may bypass the STAT3-Ser stage and act more specifically at the level of Hes3 because it has a powerful effect on inducing the nuclear translocation of Hes3.
Cells in which it operates
The effects of a particular signal transduction pathway can be very different among distinct cell types. For example, the same signal transduction pathway may promote the survival of one cell type but the maturation of another. This depends both on the nature of a cell but also on its particular state which may change over the course of its lifetime. Identifying cell types where a signal transduction pathway is operational is a first step to uncovering potentially new properties of this pathway.
The STAT3-Ser/Hes3 signaling axis has been shown to operate on various cell types. So far, research has mostly focused on stem cells and cancerous tissue and, more recently, in the function of the endocrine pancreas:
- Adult monkey (rhesus macaque) neural stem cells.[5]
- Human cancer stem cells from glioblastoma multiforme.[11]
- In a human prostate cancer cell line, STAT3-Ser was shown to promote tumorigenesis independently of STAT3-Tyr.[12]
- Chromaffin progenitor cells of the bovine adrenal medulla.[13]
- Mouse insulinoma cells (MIN6 cell line) and mouse pancreatic islet cells.[14]
- Mouse embryonic fibroblasts (MEF) during reprogramming to the induced pluripotent stem cell state.
- Human embryonic stem cells
- Mouse neural stem cells derived from induced pluripotent stem cells.[15]
Biological consequences
An individual signal transduction pathway can regulate several proteins (e.g. kinases) as well as the activation of many genes. The consequences to the properties of the cell can be, therefore, very prominent. Identifying these properties (through theoretical predictions and experimentation) sheds light on the function of the pathway and provides possible new therapeutic targets.
Activation of the notch/STAT3-Ser/Hes3 signaling axis has significant consequences to several cell types; effects have been documented both in vitro and in vivo:
- Cultured fetal and adult rodent neural stem cells: Pro-survival effects; increased yield; increased expression of sonic hedgehog protein.[2][3][5][7][8][9]
- In vivo adult rodent neural stem cells: Increase in cell number; increased expression of Sonic hedgehog (Shh) protein.[2][3][7] Delta4 administration in the adult rodent brain has also been shown to augment the effect of basic fibroblast growth factor and epidermal growth factor in promoting the proliferation of neural precursor cells in the subventricular zone and hypothalamus following ischemic stroke.[16][17]
- Cultured adult monkey neural stem cells: Pro-survival effects; increased yield; increased expression of sonic hedgehog protein.[5]
- Cultured putative glioblastoma multiforme cancer stem cells: Pro-survival effects (Hes3 knockdown by RNA interference reduces cell number).[11]
- Cultured bovine chromaffin progenitor cells: Several activators of the signaling pathway increase cell yield.[13]
- Cultured mouse insulinoma cells (MIN6 cell line): These cells can be cultured efficiently under conditions that promote the operation of the signaling pathway; Hes3 RNA interference opposes growth and the release of insulin following standard protocols that evoke insulin release from these cells.[18]
- Mice that are engineered to lack the Hes3 gene exhibit increased sensitivity to treatments that damage endocrine pancreas cells.
- Recent research implicates Hes3 in direct reprogramming of adult mouse cells to the neural stem cell state; a causative relation remains to be determined.[19]
- Hes3 and components of the Signaling Axis are regulated during critical stages of reprogramming (Mouse Embryonic Fibroblast - to - Embryonic Stem Cell reprogramming).[20]
- Mice genetically engineered to lack the Hes3 gene fail to upregulate the transcription factor Neurogenin3 during pancreatic regeneration (induced by streptozotocin treatment).[20] This is indicative of a compromised regenerative response.
Role in the adult brain
As stated above, the STAT3-Ser/Hes3 signaling axis regulates the number of neural stem cells (as well as other cell types) in culture. This prompted experiments to determine if the same pathway can also regulate the number of naturally resident (endogenous) neural stem cells in the adult rodent brain. If so, this would generate a new experimental approach to study the effects of increasing the number of endogenous neural stem cells (eNSCs). For example, would this lead to the replacement of lost cells by newly generated cells from eNSCs? Or, could this lead to the rescue of damaged neurons in models of neurodegenerative disease, since eNSCs are known to produce factors that can protect injured neurons?[21]
Various treatments that input into the STAT3-Ser/Hes3 signaling axis (Delta4, Angiopoietin 2, insulin, or a combined treatment consisting of all three factors and an inhibitor of JAK) induce the increase in numbers of endogenous neural stem cells as well as behavioral recovery in models of neurodegenerative disease. Several pieces of evidence suggest that in the adult brain, pharmacological activation of the STAT3-Ser/Hes3 signaling axis protects compromised neurons through increased neurotrophic support provided by activated neural stem cells / neural precursor cells, which can be identified by their expression of Hes3:
- These treatments increase the number of Hes3+ cells by several-fold.[2][3][8][9]
- Hes3+ cells can be isolated and placed in culture where they exhibit stem cell properties.[2][3][5][7][8]
- In culture and in vivo, Hes3+ cells express Shh, which supports the survival of certain neurons [Hes3+ cells may also express other pro-survival factors, yet unidentified].[2][3]
- The distribution of Hes3+ cells in the adult brain is widespread and can be found in close physical proximity to different types of neurons.[3]
- Diverse treatments that converge to the STAT3-Ser/Hes3 signaling axis exert similar effects in the normal brain (increase in the number of Hes3+ cells) and in the compromised brain (increase in the number of Hes3+ cells, oppose neuronal death, and improve behavioral state).[2][3][7][9]
- Macrophage migration inhibitory factor stimulates this signaling pathway and promotes the survival of neural stem cells.[10]
- Mice genetically engineered to lack the Hes3 gene exhibit differences in the amount of myelin basic protein (a protein expressed on myelinating oligodendrocytes), relative to normal mice; Hes3-lacking mice also exhibit a different regulation of this protein after oligodendrocyte damage induced by the chemical cuprizone.[22]
Implications to disease
The emerging understanding of the role of eNSCs in the adult mammalian brain suggested the relevance of these cells to disease. To address this issue, experiments were performed where the activation of eNSCs was induced in models of disease. This allowed the study of the consequences of activating eNSCs in the diseased brain. Several lines of evidence implicate the STAT3-Ser/Hes3 signaling axis in various diseases:
- Activation of the signaling pathway by Delta4 in combination with basic fibroblast growth factor (bFGF) induces motor and sensory skill improvements in adult rat models of ischemic stroke (PMCAO model).[2]
- This signaling pathway may mediate pro-survival functions of macrophage migration inhibitory factor on neural stem cells.[10]
- Activation of the signaling pathway by Delta4, Angiopoietin 2, insulin, or a combination of the three and a JAK inhibitor induces motor skill improvements in adult rat models of Parkinson's disease (6-hydroxydopamine model).[3][7]
- RNA interference (“knockdown”) of Hes3 in cultures of cells with cancer stem cell properties from patients with glioblastoma multiforme reduces cell number.[11]
- Mice lacking Hes3 exhibit increased sensitivity to particular paradigms of pancreatic islet damage, suggesting roles in diabetes.[13]
Tissue cytoarchitecture
In tissues, many different cell types interact with one another. In the brain, for example, neurons, astrocytes, and oligodendrocytes (specialized cells of the neural tissue, each with specific functions) interact with one another as well as with cells that comprise blood vessels. All these different cell types may interact with all others by the production of ligands that may activate receptors on the cell surface of other cell types. Understanding the way these different cell types interact with one another will allow to predict ways of activating eNSCs. For example, because eNSCs are found in close proximity with blood vessels, it has been hypothesized that signals (e.g., ligands) from cells comprising the blood vessel act on receptors found on the cell surface of eNSCs.
Endogenous neural stem cells are often in close physical proximity to blood vessels. Signals from blood vessels regulate their interaction with stem cells and contribute to the cytoarchitecture of the tissue. The STAT3-Ser/Hes3 signaling axis operating in Hes3+ cells is a convergence point for several of these signals (e.g. Delta4, Angiopoietin 2). Hes3, in turn, by regulating the expression of Shh and potentially other factors, can also exert an effect on blood vessels and other cells comprising their microenvironment.
References
- Artavanis-Tsakonas S, Rand MD, Lake RJ (April 1999). "Notch signaling: cell fate control and signal integration in development". Science. 284 (5415): 770–6. doi:10.1126/science.284.5415.770. PMID 10221902.
- Androutsellis-Theotokis A, Leker RR, Soldner F, et al. (August 2006). "Notch signalling regulates stem cell numbers in vitro and in vivo" (PDF). Nature. 442 (7104): 823–6. doi:10.1038/nature04940. PMID 16799564.
- Androutsellis-Theotokis A, Rueger MA, Park DM, et al. (August 2009). "Targeting neural precursors in the adult brain rescues injured dopamine neurons". Proc. Natl. Acad. Sci. U.S.A. 106 (32): 13570–5. doi:10.1073/pnas.0905125106. PMC 2714762. PMID 19628689.
- Kageyama R, Ohtsuka T, Kobayashi T (June 2008). "Roles of Hes genes in neural development". Dev. Growth Differ. 50 Suppl 1: S97–103. doi:10.1111/j.1440-169X.2008.00993.x. PMID 18430159.
- Androutsellis-Theotokis A, Rueger MA, Park DM, et al. (2010). "Angiogenic factors stimulate growth of adult neural stem cells". PLoS ONE. 5 (2): e9414. doi:10.1371/journal.pone.0009414. PMC 2829079. PMID 20195471.
- Levy DE, Darnell JE (September 2002). "Stats: transcriptional control and biological impact". Nat. Rev. Mol. Cell Biol. 3 (9): 651–62. doi:10.1038/nrm909. PMID 12209125.
- Androutsellis-Theotokis A, Rueger MA, Mkhikian H, Korb E, McKay RD (2008). "Signaling pathways controlling neural stem cells slow progressive brain disease". Cold Spring Harb. Symp. Quant. Biol. 73: 403–10. doi:10.1101/sqb.2008.73.018. PMID 19022746.
- Androutsellis-Theotokis A, Walbridge S, Park DM, Lonser RR, McKay RD (2010). "Cholera toxin regulates a signaling pathway critical for the expansion of neural stem cell cultures from the fetal and adult rodent brains". PLoS ONE. 5 (5): e10841. doi:10.1371/journal.pone.0010841. PMC 2877108. PMID 20520777.
- Masjkur J, Rueger MA, Bornstein SR, McKay R, Androutsellis-Theotokis A (November 2012). "Neurovascular signals suggest a propagation mechanism for endogenous stem cell activation along blood vessels". CNS Neurol Disord Drug Targets. 11 (7): 805–17. doi:10.2174/1871527311201070805. PMC 3580829. PMID 23131162.
- Ohta S, Misawa A, Fukaya R, et al. (July 2012). "Macrophage migration inhibitory factor (MIF) promotes cell survival and proliferation of neural stem/progenitor cells". J. Cell Sci. 125 (Pt 13): 3210–20. doi:10.1242/jcs.102210. PMID 22454509.
- Park DM, Jung J, Masjkur J, et al. (2013). "Hes3 regulates cell number in cultures from glioblastoma multiforme with stem cell characteristics". Sci Rep. 3: 1095. doi:10.1038/srep01095. PMC 3566603. PMID 23393614.
- Qin HR, Kim HJ, Kim JY, et al. (October 2008). "Activation of signal transducer and activator of transcription 3 through a phosphomimetic serine 727 promotes prostate tumorigenesis independent of tyrosine 705 phosphorylation". Cancer Res. 68 (19): 7736–41. doi:10.1158/0008-5472.CAN-08-1125. PMC 2859454. PMID 18829527.
- Masjkur J, et al. (July 2014). "A defined, controlled culture system for primary bovine chromaffin progenitors reveals novel biomarkers and modulators". Stem Cells Transl Med. 3 (7): 801–8. doi:10.5966/sctm.2013-0211. PMC 4073824. PMID 24855275.
- Masjkur J, et al. (December 2014). "Hes3 is expressed in the adult pancreatic islet and regulates gene expression, cell growth, and insulin release". J Biol Chem. 289 (51): 35503–16. doi:10.1074/jbc.M114.590687. PMC 4271235. PMID 25371201.
- Salewski RP, et al. (February 2013). "The generation of definitive neural stem cells from PiggyBac transposon-induced pluripotent stem cells can be enhanced by induction of the NOTCH signaling pathway". Stem Cells Dev. 22 (3): 383–96. doi:10.1089/scd.2012.0218. PMC 3549637. PMID 22889305.
- Oya S, Yoshikawa G, Takai K, et al. (May 2008). "Region-specific proliferative response of neural progenitors to exogenous stimulation by growth factors following ischemia". NeuroReport. 19 (8): 805–9. doi:10.1097/WNR.0b013e3282ff8641. PMID 18463491.
- Wang L, Chopp M, Zhang RL, et al. (February 2009). "The Notch pathway mediates expansion of a progenitor pool and neuronal differentiation in adult neural progenitor cells after stroke". Neuroscience. 158 (4): 1356–63. doi:10.1016/j.neuroscience.2008.10.064. PMC 2757073. PMID 19059466.
- Masjkur J, Poser SW, Nikolakopoulou P, Chrousos G, McKay RD, Bornstein SR, Jones PM, Androutsellis-Theotokis A (2016). "Endocrine Pancreas Development and Regeneration: Noncanonical Ideas From Neural Stem Cell Biology". Diabetes. 65: 314–30. doi:10.2337/db15-1099. PMID 26798118.
- Cassady JP, et al. (December 2014). "Direct lineage conversion of adult mouse liver cells and B lymphocytes to neural stem cells". Stem Cell Reports. 3 (6): 948–56. doi:10.1016/j.stemcr.2014.10.001. PMC 4264067. PMID 25454632.
- Poser SW, Chenoweth JG, Colantuoni C, Masjkur J, Chrousos G, Bornstein SR, McKay RD, and Androutsellis-Theotokis A (2015). "Reprogramming, behind-the-scenes: Non-canonical neural stem cell signaling pathways reveal new, unseen regulators of tissue plasticity with therapeutic implications". Stem Cells Transl Med. 4: 1251–7. doi:10.5966/sctm.2015-0105. PMC 4622411. PMID 26371344.
- Kittappa R, Bornstein SR, Androutsellis-Theotokis A (December 2012). "The role of eNSCs in neurodegenerative disease". Mol. Neurobiol. 46 (3): 555–62. doi:10.1007/s12035-012-8303-8. PMID 22821143.
- Toutouna L, Nikolakopoulou P, Poser SW, Masjkur J, Arps-Forker C, Troullinaki M, Grossklaus S, Bosak V, Friedrich U, Ziemssen T, Bornstein SR, Chavakis T, Androutsellis-Theotokis A (2016). "Hes3 expression in the adult mouse brain is regulated during demyelination and remyelination". Brain Research. 1642: 124–30. doi:10.1016/j.brainres.2016.03.014. PMID 27018293.