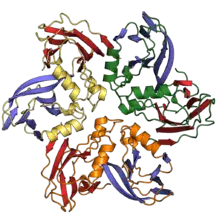
Jelly roll fold
The jelly roll or Swiss roll fold is a protein fold or supersecondary structure composed of eight beta strands arranged in two four-stranded sheets. The name of the structure was introduced by Jane S. Richardson in 1981, reflecting its resemblance to the jelly or Swiss roll cake.[2] The fold is an elaboration on the Greek key motif and is sometimes considered a form of beta barrel. It is very common in viral proteins, particularly viral capsid proteins.[3][4] Taken together, the jelly roll and Greek key structures comprise around 30% of the all-beta proteins annotated in the Structural Classification of Proteins (SCOP) database.[5]
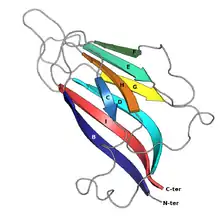
Structure
The basic jelly roll structure consists of eight beta strands arranged in two four-stranded antiparallel beta sheets which pack together across a hydrophobic interface. The strands are traditionally labeled B through I for the historical reason that the first solved structure, of a jelly roll capsid protein from the tomato bushy stunt virus, had an additional strand A outside the fold's common core.[6][7] The sheets are composed of strands BIDG and CHEF, folded such that strand B packs opposite strand C, I opposite H, etc.[4][8]
Viral proteins
A large number of viruses build their exterior capsids from proteins containing either a single or a double jelly roll fold. This shared capsid architecture is thought to reflect ancient evolutionary relationships, possibly dating to before the last universal common ancestor (LUCA) of cellular life.[8][9][10] Other viral lineages use evolutionarily unrelated proteins to build their enclosed capsids, which likely evolved independently at least twice[9][11] and possibly many times, with links to proteins of cellular origin.[12]
Single jelly roll capsid proteins
Single jelly roll capsid (JRC) proteins are found in at least sixteen distinct viral families, mostly with icosahedral capsid structures and including both RNA viruses and DNA viruses.[13] Many viruses with single jelly roll capsids are positive-sense single-stranded RNA viruses. Two groups of double-stranded DNA viruses with single-JRC capsids are the Papillomaviridae and Polyomaviridae, both of which have fairly small capsids; in these viruses, the architecture of the assembled capsid orients the axis of the jelly roll parallel or "horizontally" relative to the capsid surface.[11] A large-scale analysis of viral capsid components suggested that the single horizontal jelly roll is the most common fold among capsid proteins, accounting for about 28% of known examples.[12]
Another group of viruses uses single jelly roll proteins in their capsids, but in the vertical rather than horizontal orientation. These viruses are evolutionarily related to the large group of double jelly-roll viruses known as the PRD1-adenovirus viral lineage, with similar capsid architecture realized through assembly of two distinct single jelly-roll major capsid proteins expressed from distinct genes.[14][15] These single vertical jelly-roll viruses comprise the taxon Helvetiavirae.[16] Known viruses with vertical single jelly roll capsids infect extremophilic prokaryotes.[14][12]
Double jelly roll proteins

Double jelly roll capsid proteins consist of two single jelly roll folds connected by a short linker region. They are found exclusively in double-stranded DNA viruses of at least nine different viral families, including viruses that infect all domains of life, and spanning a large capsid size range.[4][11] In the double jelly roll capsid architecture, the jelly roll axis is oriented perpendicular or "vertically" relative to the capsid surface.[18]
Double jelly roll proteins are believed to have evolved from single jelly roll proteins by gene duplication.[11][18] It is likely that vertical single jelly roll viruses represent a transitional form, and that the vertical and horizontal jelly roll capsid proteins have independent evolutionary origins from ancestral cellular proteins.[12] The degree of structural similarity among double-jelly-roll virus capsids has led to the conclusion that these viruses likely have a common evolutionary origin despite their diversity in size and in host range; this has become known as the PRD1-adenovirus lineage (Bamfordvirae).[18][16][19][20] Many members of this group have been identified through metagenomics and in some cases have few to no other viral genes in common.[12][21] Although most members of this group have icosahedral capsid geometry, a few families such as the Poxviridae and Ascoviridae have oval or brick-shaped mature virions; poxviruses such as Vaccinia undergo dramatic conformational changes mediated by highly derived double jelly roll proteins during maturation and likely derive from an icosahedral ancestor.[11][22] Shared double-jelly-roll capsid proteins, along with other homologous proteins, have also been cited in support of the proposed order Megavirales containing the nucleocytoplasmic large DNA viruses (NCLDV).[23]
Double jelly roll proteins have not been observed in cellular proteins; they appear to be unique to viruses.[11] For this reason, detecting clear homology to double jelly roll proteins in the sequences of polinton/Maverick transposable elements widespread in eukaryotic genomes is considered evidence of these genetic elements' close evolutionary relationship to viruses.[24]
Non-capsid proteins
Single jelly rolls also occur in non-capsid viral proteins, including minor components of the assembled virion as well as non-virion proteins such as polyhedrin.[11]
Cellular proteins
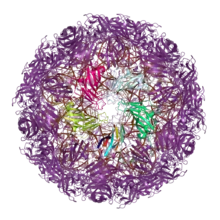
While double jelly rolls are not found in proteins of cellular origin, single jelly rolls do occur.[11][12] One such class of cellular proteins is the nucleoplasmins, which serve as molecular chaperone proteins for histone assembly into nucleosomes. The N-terminal domain of nucleoplasmins possesses a single jelly roll fold and assembled into a pentamer.[25] Similar structures have since been reported in additional groups of chromatin remodeling proteins.[26] Jelly roll motifs with identical beta-sheet connectivity are also found in tumor necrosis factor ligands[27] and proteins from the bacterium Yersinia pseudotuberculosis that belong to a class of viral and bacterial proteins known as superantigens.[28][29]
More broadly, the members of the extremely diverse cupin superfamily are also often described as jelly rolls; though the common core of the cupin domain structure contains only six beta strands, many cupins have eight.[30] Examples include the non-heme dioxygenase enzymes[31][32] and JmjC-family histone demethylases.[33][34]
Evolution
Comparative studies of proteins classified as jelly roll and Greek key structures suggest that the Greek key proteins evolved significantly earlier than their more topologically complex jelly roll counterparts.[5] Structural bioinformatics studies comparing virus capsid jelly-roll proteins to other proteins of known structure indicates that the capsid proteins form a well-separated cluster, suggesting that they are subject to a distinctive set of evolutionary constraints.[4] One of the most notable features of viral capsid jelly roll proteins is their ability to form oligomers in a repeated tiling pattern to produce a closed protein shell; the cellular proteins that are most similar in fold and topology are mostly also oligomers.[4] It has been proposed that viral jelly-roll capsid proteins have evolved from cellular jelly-roll proteins, potentially on several independent occasions, at the earliest stages of cellular evolution.[12]
History and nomenclature
The name "jelly roll" was first used for the structure composed of an elaboration on the Greek key motif by Jane S. Richardson in 1981 and was intended to reflect the structure's resemblance to a jelly or Swiss roll cake.[2] The structure has been given a variety of descriptive names, including a wedge, beta barrel, and beta roll. The edges of the two sheets do not meet to form regular hydrogen bonding patterns, and so it is often not considered to be a true beta barrel,[3] though the term is in common use in describing viral capsid architecture.[14][15] Cellular proteins containing jelly roll-like structures may be described as a cupin fold, a JmjC fold, or a double-stranded beta helix.[32]
References
- Larson, Steven B.; Day, John S.; McPherson, Alexander (29 August 2014). "Satellite tobacco mosaic virus refined to 1.4 Å resolution". Acta Crystallographica Section D. 70 (9): 2316–2330. doi:10.1107/S1399004714013789. PMC 4157444. PMID 25195746.
- Richardson, JS (1981). "The anatomy and taxonomy of protein structure". Advances in Protein Chemistry Volume 34. Advances in Protein Chemistry. 34. pp. 167–339. doi:10.1016/S0065-3233(08)60520-3. ISBN 9780120342341. PMID 7020376.
- Chelvanayagam, Gareth; Heringa, Jaap; Argos, Patrick (November 1992). "Anatomy and evolution of proteins displaying the viral capsid jellyroll topology". Journal of Molecular Biology. 228 (1): 220–242. doi:10.1016/0022-2836(92)90502-B. PMID 1447783.
- Cheng, Shanshan; Brooks, Charles L.; Livesay, Dennis R. (7 February 2013). "Viral Capsid Proteins Are Segregated in Structural Fold Space". PLoS Computational Biology. 9 (2): e1002905. Bibcode:2013PLSCB...9E2905C. doi:10.1371/journal.pcbi.1002905. PMC 3567143. PMID 23408879.
- Edwards, Hannah; Abeln, Sanne; Deane, Charlotte M.; Orengo, Christine A. (14 November 2013). "Exploring Fold Space Preferences of New-born and Ancient Protein Superfamilies". PLoS Computational Biology. 9 (11): e1003325. doi:10.1371/journal.pcbi.1003325. PMC 3828129. PMID 24244135.
- Harrison, S. C.; Olson, A. J.; Schutt, C. E.; Winkler, F. K.; Bricogne, G. (23 November 1978). "Tomato bushy stunt virus at 2.9 Å resolution". Nature. 276 (5686): 368–373. Bibcode:1978Natur.276..368H. doi:10.1038/276368a0. PMID 19711552.
- Rossmann, Michael G.; Abad-Zapatero, Celerino; Murthy, Mathur R.N.; Liljas, Lars; Jones, T. Alwyn; Strandberg, Bror (April 1983). "Structural comparisons of some small spherical plant viruses". Journal of Molecular Biology. 165 (4): 711–736. doi:10.1016/S0022-2836(83)80276-9. PMID 6854630.
- Benson, Stacy D.; Bamford, Jaana K.H.; Bamford, Dennis H.; Burnett, Roger M. (December 2004). "Does Common Architecture Reveal a Viral Lineage Spanning All Three Domains of Life?". Molecular Cell. 16 (5): 673–685. doi:10.1016/j.molcel.2004.11.016. PMID 15574324.
- Forterre, Patrick; Prangishvili, David (September 2009). "The origin of viruses". Research in Microbiology. 160 (7): 466–472. doi:10.1016/j.resmic.2009.07.008. PMID 19647075.
- Holmes, E. C. (30 March 2011). "What Does Virus Evolution Tell Us about Virus Origins?". Journal of Virology. 85 (11): 5247–5251. doi:10.1128/JVI.02203-10. PMC 3094976. PMID 21450811.
- Krupovic, Mart; Bamford, Dennis H (August 2011). "Double-stranded DNA viruses: 20 families and only five different architectural principles for virion assembly". Current Opinion in Virology. 1 (2): 118–124. doi:10.1016/j.coviro.2011.06.001. PMID 22440622.
- Krupovic, Mart; Koonin, Eugene V. (21 March 2017). "Multiple origins of viral capsid proteins from cellular ancestors". Proceedings of the National Academy of Sciences. 114 (12): E2401–E2410. doi:10.1073/pnas.1621061114.
- Krupovic M (2013). "Networks of evolutionary interactions underlying the polyphyletic origin of ssDNA viruses". Current Opinion in Virology. 3 (5): 578–586. doi:10.1016/j.coviro.2013.06.010. PMID 23850154.
- Gil-Carton, David; Jaakkola, Salla T.; Charro, Diego; Peralta, Bibiana; Castaño-Díez, Daniel; Oksanen, Hanna M.; Bamford, Dennis H.; Abrescia, Nicola G.A. (October 2015). "Insight into the Assembly of Viruses with Vertical Single β-barrel Major Capsid Proteins". Structure. 23 (10): 1866–1877. doi:10.1016/j.str.2015.07.015.
- Santos-Pérez, Isaac; Charro, Diego; Gil-Carton, David; Azkargorta, Mikel; Elortza, Felix; Bamford, Dennis H.; Oksanen, Hanna M.; Abrescia, Nicola G. A. (December 2019). "Structural basis for assembly of vertical single β-barrel viruses". Nature Communications. 10 (1): 1184. doi:10.1038/s41467-019-08927-2.
- Koonin EV, Dolja VV, Krupovic M, Varsani A, Wolf YI, Yutin N, Zerbini M, Kuhn JH (October 2019). "Create a megataxonomic framework, filling all principal taxonomic ranks, for DNA viruses encoding vertical jelly roll-type major capsid proteins". ICTV Proposal (Taxoprop): 2019.003G. doi:10.13140/RG.2.2.14886.47684.
- Abrescia, Nicola G.A.; Grimes, Jonathan M.; Kivelä, Hanna M.; Assenberg, Rene; Sutton, Geoff C.; Butcher, Sarah J.; Bamford, Jaana K.H.; Bamford, Dennis H.; Stuart, David I. (September 2008). "Insights into Virus Evolution and Membrane Biogenesis from the Structure of the Marine Lipid-Containing Bacteriophage PM2". Molecular Cell. 31 (5): 749–761. doi:10.1016/j.molcel.2008.06.026. PMID 18775333.
- Krupovič, Mart; Bamford, Dennis H. (December 2008). "Virus evolution: how far does the double β-barrel viral lineage extend?". Nature Reviews Microbiology. 6 (12): 941–948. doi:10.1038/nrmicro2033. PMID 19008892.
- Koonin, Eugene V.; Dolja, Valerian V.; Krupovic, Mart; Varsani, Arvind; Wolf, Yuri I.; Yutin, Natalya; Zerbini, F. Murilo; Kuhn, Jens H. (4 March 2020). "Global Organization and Proposed Megataxonomy of the Virus World". Microbiology and Molecular Biology Reviews. 84 (2): e00061–19, /mmbr/84/2/MMBR.00061–19.atom. doi:10.1128/MMBR.00061-19.
- Walker, Peter J.; Siddell, Stuart G.; Lefkowitz, Elliot J.; Mushegian, Arcady R.; Adriaenssens, Evelien M.; Dempsey, Donald M.; Dutilh, Bas E.; Harrach, Balázs; Harrison, Robert L.; Hendrickson, R. Curtis; Junglen, Sandra; Knowles, Nick J.; Kropinski, Andrew M.; Krupovic, Mart; Kuhn, Jens H.; Nibert, Max; Orton, Richard J.; Rubino, Luisa; Sabanadzovic, Sead; Simmonds, Peter; Smith, Donald B.; Varsani, Arvind; Zerbini, Francisco Murilo; Davison, Andrew J. (November 2020). "Changes to virus taxonomy and the Statutes ratified by the International Committee on Taxonomy of Viruses (2020)". Archives of Virology. 165 (11): 2737–2748. doi:10.1007/s00705-020-04752-x.
- Yutin, Natalya; Bäckström, Disa; Ettema, Thijs J. G.; Krupovic, Mart; Koonin, Eugene V. (10 April 2018). "Vast diversity of prokaryotic virus genomes encoding double jelly-roll major capsid proteins uncovered by genomic and metagenomic sequence analysis". Virology Journal. 15 (1): 67. doi:10.1186/s12985-018-0974-y.
- Bahar, Mohammad W.; Graham, Stephen C.; Stuart, David I.; Grimes, Jonathan M. (July 2011). "Insights into the Evolution of a Complex Virus from the Crystal Structure of Vaccinia Virus D13". Structure. 19 (7): 1011–1020. doi:10.1016/j.str.2011.03.023. PMC 3136756. PMID 21742267.
- Colson, Philippe; De Lamballerie, Xavier; Yutin, Natalya; Asgari, Sassan; Bigot, Yves; Bideshi, Dennis K.; Cheng, Xiao-Wen; Federici, Brian A.; Van Etten, James L.; Koonin, Eugene V.; La Scola, Bernard; Raoult, Didier (29 June 2013). ""Megavirales", a proposed new order for eukaryotic nucleocytoplasmic large DNA viruses". Archives of Virology. 158 (12): 2517–2521. doi:10.1007/s00705-013-1768-6. PMC 4066373. PMID 23812617.
- Krupovic, Mart; Bamford, Dennis H; Koonin, Eugene V (2014). "Conservation of major and minor jelly-roll capsid proteins in Polinton (Maverick) transposons suggests that they are bona fide viruses". Biology Direct. 9 (1): 6. doi:10.1186/1745-6150-9-6. PMC 4028283. PMID 24773695.
- Dutta, Shuchismita; Akey, Ildikó V.; Dingwall, Colin; Hartman, Kari L.; Laue, Tom; Nolte, Robert T.; Head, James F.; Akey, Christopher W. (October 2001). "The Crystal Structure of Nucleoplasmin-Core". Molecular Cell. 8 (4): 841–853. doi:10.1016/S1097-2765(01)00354-9. PMID 11684019.
- Edlich-Muth, Christian; Artero, Jean-Baptiste; Callow, Phil; Przewloka, Marcin R.; Watson, Aleksandra A.; Zhang, Wei; Glover, David M.; Debski, Janusz; Dadlez, Michal; Round, Adam R.; Forsyth, V. Trevor; Laue, Ernest D. (May 2015). "The Pentameric Nucleoplasmin Fold Is Present in Drosophila FKBP39 and a Large Number of Chromatin-Related Proteins". Journal of Molecular Biology. 427 (10): 1949–1963. doi:10.1016/j.jmb.2015.03.010. PMC 4414354. PMID 25813344.
- Bodmer, Jean-Luc; Schneider, Pascal; Tschopp, Jürg (January 2002). "The molecular architecture of the TNF superfamily" (PDF). Trends in Biochemical Sciences. 27 (1): 19–26. doi:10.1016/S0968-0004(01)01995-8. PMID 11796220.
- Donadini, Roberta; Liew, Chu Wai; Kwan, Ann H.Y.; Mackay, Joel P.; Fields, Barry A. (March 2004). "Crystal and Solution Structures of a Superantigen from Yersinia pseudotuberculosis Reveal a Jelly-Roll Fold". Structure. 12 (1): 145–156. doi:10.1016/j.str.2003.12.002. PMID 14725774.
- Fraser, John D.; Proft, Thomas (October 2008). "The bacterial superantigen and superantigen-like proteins". Immunological Reviews. 225 (1): 226–243. doi:10.1111/j.1600-065X.2008.00681.x. PMID 18837785.
- Khuri, S; Bakker, FT; Dunwell, JM (April 2001). "Phylogeny, function, and evolution of the cupins, a structurally conserved, functionally diverse superfamily of proteins". Molecular Biology and Evolution. 18 (4): 593–605. doi:10.1093/oxfordjournals.molbev.a003840. PMID 11264412.
- Ozer, Abdullah; Bruick, Richard K (March 2007). "Non-heme dioxygenases: cellular sensors and regulators jelly rolled into one?". Nature Chemical Biology. 3 (3): 144–153. doi:10.1038/nchembio863. PMID 17301803.
- Aik, WeiShen; McDonough, Michael A; Thalhammer, Armin; Chowdhury, Rasheduzzaman; Schofield, Christopher J (December 2012). "Role of the jelly-roll fold in substrate binding by 2-oxoglutarate oxygenases". Current Opinion in Structural Biology. 22 (6): 691–700. doi:10.1016/j.sbi.2012.10.001. PMID 23142576.
- Chen, Zhongzhou; Zang, Jianye; Whetstine, Johnathan; Hong, Xia; Davrazou, Foteini; Kutateladze, Tatiana G.; Simpson, Michael; Mao, Qilong; Pan, Cheol-Ho; Dai, Shaodong; Hagman, James; Hansen, Kirk; Shi, Yang; Zhang, Gongyi (May 2006). "Structural Insights into Histone Demethylation by JMJD2 Family Members". Cell. 125 (4): 691–702. doi:10.1016/j.cell.2006.04.024. PMID 16677698.
- Klose, Robert J.; Zhang, Yi (7 March 2007). "Regulation of histone methylation by demethylimination and demethylation". Nature Reviews Molecular Cell Biology. 8 (4): 307–318. doi:10.1038/nrm2143. PMID 17342184.
External links
- Antiparallel β Domains, a section from Anatomy and Taxonomy of Protein Structure by Jane S. Richardson
- The Jelly Roll of Life by Jacqueline Humphries at Small Things Considered, a blog sponsored by the American Society for Microbiology