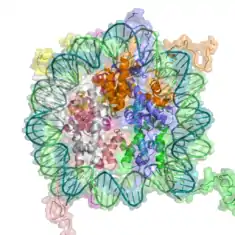
Chromatin
Chromatin is a complex of DNA and protein found in eukaryotic cells.[1] Its primary function is packaging long DNA molecules into more compact, denser structures. This prevents the strands from becoming tangled and also plays important roles in reinforcing the DNA during cell division, preventing DNA damage, and regulating gene expression and DNA replication. During mitosis and meiosis, chromatin facilitates proper segregation of the chromosomes in anaphase; the characteristic shapes of chromosomes visible during this stage are the result of DNA being coiled into highly condensed chromatin.
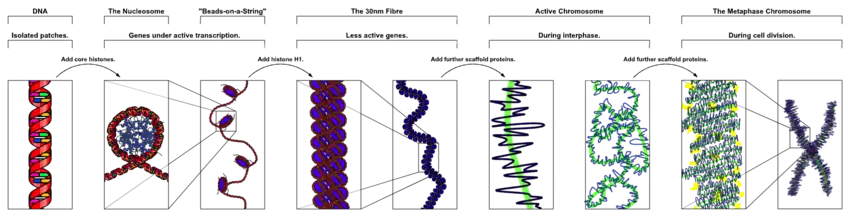
The primary protein components of chromatin are histones, which bind to DNA and function as "anchors" around which the strands are wound. In general, there are three levels of chromatin organization:
- DNA wraps around histone proteins, forming nucleosomes and the so-called beads on a string structure (euchromatin).
- Multiple histones wrap into a 30-nanometer fibre consisting of nucleosome arrays in their most compact form (heterochromatin).[lower-alpha 1]
- Higher-level DNA supercoiling of the 30-nm fiber produces the metaphase chromosome (during mitosis and meiosis).
Many organisms, however, do not follow this organization scheme. For example, spermatozoa and avian red blood cells have more tightly packed chromatin than most eukaryotic cells, and trypanosomatid protozoa do not condense their chromatin into visible chromosomes at all. Prokaryotic cells have entirely different structures for organizing their DNA (the prokaryotic chromosome equivalent is called a genophore and is localized within the nucleoid region).
The overall structure of the chromatin network further depends on the stage of the cell cycle. During interphase, the chromatin is structurally loose to allow access to RNA and DNA polymerases that transcribe and replicate the DNA. The local structure of chromatin during interphase depends on the specific genes present in the DNA. Regions of DNA containing genes which are actively transcribed ("turned on") are less tightly compacted and closely associated with RNA polymerases in a structure known as euchromatin, while regions containing inactive genes ("turned off") are generally more condensed and associated with structural proteins in heterochromatin.[3] Epigenetic modification of the structural proteins in chromatin via methylation and acetylation also alters local chromatin structure and therefore gene expression. The structure of chromatin networks is currently poorly understood and remains an active area of research in molecular biology.
Dynamic chromatin structure and hierarchy
Chromatin undergoes various structural changes during a cell cycle. Histone proteins are the basic packers and arrangers of chromatin and can be modified by various post-translational modifications to alter chromatin packing (histone modification). Most modifications occur on histone tails. The consequences in terms of chromatin accessibility and compaction depend both on the modified amino acid and the type of modification. For example, histone acetylation results in loosening and increased accessibility of chromatin for replication and transcription. Lysine trimethylation can either lead to increased transcriptional activity (trimethylation of histone H3 lysine 4) or transcriptional repression and chromatin compaction (trimethylation of histone H3 lysine 9 or 27). Several studies suggested that different modifications could occur simultaneously. For example, it was proposed that a bivalent structure (with trimethylation of both lysine 4 and 27 on histone H3) is involved in early mammalian development.[4]
Polycomb-group proteins play a role in regulating genes through modulation of chromatin structure.[5]
For additional information, see Histone modifications in chromatin regulation and RNA polymerase control by chromatin structure.
DNA structure

In nature, DNA can form three structures, A-, B-, and Z-DNA. A- and B-DNA are very similar, forming right-handed helices, whereas Z-DNA is a left-handed helix with a zig-zag phosphate backbone. Z-DNA is thought to play a specific role in chromatin structure and transcription because of the properties of the junction between B- and Z-DNA.
At the junction of B- and Z-DNA, one pair of bases is flipped out from normal bonding. These play a dual role of a site of recognition by many proteins and as a sink for torsional stress from RNA polymerase or nucleosome binding.
Nucleosomes and beads-on-a-string
- Main articles: Nucleosome, Chromatosome and Histone
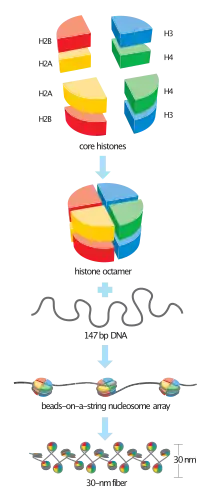
The basic repeat element of chromatin is the nucleosome, interconnected by sections of linker DNA, a far shorter arrangement than pure DNA in solution.
In addition to core histones, a linker histone H1 exists that contacts the exit/entry of the DNA strand on the nucleosome. The nucleosome core particle, together with histone H1, is known as a chromatosome. Nucleosomes, with about 20 to 60 base pairs of linker DNA, can form, under non-physiological conditions, an approximately 10 nm beads on a string fibre.
The nucleosomes bind DNA non-specifically, as required by their function in general DNA packaging. There are, however, large DNA sequence preferences that govern nucleosome positioning. This is due primarily to the varying physical properties of different DNA sequences: For instance, adenine (A), and thymine (T) are more favorably compressed into the inner minor grooves. This means nucleosomes can bind preferentially at one position approximately every 10 base pairs (the helical repeat of DNA)- where the DNA is rotated to maximise the number of A and T bases that will lie in the inner minor groove. (See nucleic acid structure.)
30-nanometer chromatin fiber

Left: 1 start helix "solenoid" structure.
Right: 2 start loose helix structure.
Note: the histones are omitted in this diagram - only the DNA is shown.
With addition of H1, the beads-on-a-string structure in turn coils into a 30 nm diameter helical structure known as the 30 nm fibre or filament. The precise structure of the chromatin fiber in the cell is not known in detail.[6]
This level of chromatin structure is thought to be the form of heterochromatin, which contains mostly transcriptionally silent genes. Electron microscopy studies have demonstrated that the 30 nm fiber is highly dynamic such that it unfolds into a 10 nm fiber beads-on-a-string structure when transversed by an RNA polymerase engaged in transcription.
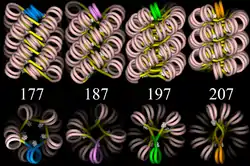
Linker DNA in yellow and nucleosomal DNA in pink.
The existing models commonly accept that the nucleosomes lie perpendicular to the axis of the fibre, with linker histones arranged internally. A stable 30 nm fibre relies on the regular positioning of nucleosomes along DNA. Linker DNA is relatively resistant to bending and rotation. This makes the length of linker DNA critical to the stability of the fibre, requiring nucleosomes to be separated by lengths that permit rotation and folding into the required orientation without excessive stress to the DNA. In this view, different lengths of the linker DNA should produce different folding topologies of the chromatin fiber. Recent theoretical work, based on electron-microscopy images[7] of reconstituted fibers supports this view.[8]
Spatial organization of chromatin in the cell nucleus
The spatial arrangement of the chromatin within the nucleus is not random - specific regions of the chromatin can be found in certain territories. Territories are, for example, the lamina-associated domains (LADs), and the topologically associating domains (TADs), which are bound together by protein complexes.[9] Currently, polymer models such as the Strings & Binders Switch (SBS) model[10] and the Dynamic Loop (DL) model[11] are used to describe the folding of chromatin within the nucleus.
Cell-cycle dependent structural organization


- Interphase: The structure of chromatin during interphase of mitosis is optimized to allow simple access of transcription and DNA repair factors to the DNA while compacting the DNA into the nucleus. The structure varies depending on the access required to the DNA. Genes that require regular access by RNA polymerase require the looser structure provided by euchromatin.
- Metaphase: The metaphase structure of chromatin differs vastly to that of interphase. It is optimised for physical strength and manageability, forming the classic chromosome structure seen in karyotypes. The structure of the condensed chromatin is thought to be loops of 30 nm fibre to a central scaffold of proteins. It is, however, not well-characterised. Chromosome scaffolds play an important role to hold the chromatin into compact chromosomes. Loops of 30 nm structure further condense with scaffold, into higher order structures.[12] Chromosome scaffolds are made of proteins including condensin, type IIA topoisomerase and kinesin family member 4 (KIF4).[13] The physical strength of chromatin is vital for this stage of division to prevent shear damage to the DNA as the daughter chromosomes are separated. To maximise strength the composition of the chromatin changes as it approaches the centromere, primarily through alternative histone H1 analogues. During mitosis, although most of the chromatin is tightly compacted, there are small regions that are not as tightly compacted. These regions often correspond to promoter regions of genes that were active in that cell type prior to chromatin formation. The lack of compaction of these regions is called bookmarking, which is an epigenetic mechanism believed to be important for transmitting to daughter cells the "memory" of which genes were active prior to entry into mitosis.[14] This bookmarking mechanism is needed to help transmit this memory because transcription ceases during mitosis.
Chromatin and bursts of transcription
Chromatin and its interaction with enzymes has been researched, and a conclusion being made is that it is relevant and an important factor in gene expression. Vincent G. Allfrey, a professor at Rockefeller University, stated that RNA synthesis is related to histone acetylation.[15] The lysine amino acid attached to the end of the histones is positively charged. The acetylation of these tails would make the chromatin ends neutral, allowing for DNA access.
When the chromatin decondenses, the DNA is open to entry of molecular machinery. Fluctuations between open and closed chromatin may contribute to the discontinuity of transcription, or transcriptional bursting. Other factors are probably involved, such as the association and dissociation of transcription factor complexes with chromatin. The phenomenon, as opposed to simple probabilistic models of transcription, can account for the high variability in gene expression occurring between cells in isogenic populations.[16]
Alternative chromatin organizations
During metazoan spermiogenesis, the spermatid's chromatin is remodeled into a more spaced-packaged, widened, almost crystal-like structure. This process is associated with the cessation of transcription and involves nuclear protein exchange. The histones are mostly displaced, and replaced by protamines (small, arginine-rich proteins).[17] It is proposed that in yeast, regions devoid of histones become very fragile after transcription; HMO1, an HMG-box protein, helps in stabilizing nucleosomes-free chromatin.[18][19]
Chromatin and DNA repair
The packaging of eukaryotic DNA into chromatin presents a barrier to all DNA-based processes that require recruitment of enzymes to their sites of action. To allow the critical cellular process of DNA repair, the chromatin must be remodeled. In eukaryotes, ATP-dependent chromatin remodeling complexes and histone-modifying enzymes are two predominant factors employed to accomplish this remodeling process.[20]
Chromatin relaxation occurs rapidly at the site of a DNA damage.[21] This process is initiated by PARP1 protein that starts to appear at DNA damage in less than a second, with half maximum accumulation within 1.6 seconds after the damage occurs.[22] Next the chromatin remodeler Alc1 quickly attaches to the product of PARP1, and completes arrival at the DNA damage within 10 seconds of the damage.[21] About half of the maximum chromatin relaxation, presumably due to action of Alc1, occurs by 10 seconds.[21] This then allows recruitment of the DNA repair enzyme MRE11, to initiate DNA repair, within 13 seconds.[22]
γH2AX, the phosphorylated form of H2AX is also involved in the early steps leading to chromatin decondensation after DNA damage occurrence. The histone variant H2AX constitutes about 10% of the H2A histones in human chromatin.[23] γH2AX (H2AX phosphorylated on serine 139) can be detected as soon as 20 seconds after irradiation of cells (with DNA double-strand break formation), and half maximum accumulation of γH2AX occurs in one minute.[23] The extent of chromatin with phosphorylated γH2AX is about two million base pairs at the site of a DNA double-strand break.[23] γH2AX does not, itself, cause chromatin decondensation, but within 30 seconds of irradiation, RNF8 protein can be detected in association with γH2AX.[24] RNF8 mediates extensive chromatin decondensation, through its subsequent interaction with CHD4,[25] a component of the nucleosome remodeling and deacetylase complex NuRD.
After undergoing relaxation subsequent to DNA damage, followed by DNA repair, chromatin recovers to a compaction state close to its pre-damage level after about 20 min.[21]
Methods to investigate chromatin
- ChIP-seq (Chromatin immunoprecipitation sequencing), aimed against different histone modifications, can be used to identify chromatin states throughout the genome. Different modifications have been linked to various states of chromatin.
- DNase-seq (DNase I hypersensitive sites Sequencing) uses the sensitivity of accessible regions in the genome to the DNase I enzyme to map open or accessible regions in the genome.
- FAIRE-seq (Formaldehyde-Assisted Isolation of Regulatory Elements sequencing) uses the chemical properties of protein-bound DNA in a two-phase separation method to extract nucleosome depleted regions from the genome.[26]
- ATAC-seq (Assay for Transposable Accessible Chromatin sequencing) uses the Tn5 transposase to integrate (synthetic) transposons into accessible regions of the genome consequentially highlighting the localisation of nucleosomes and transcription factors across the genome.
- DNA footprinting is a method aimed at identifying protein-bound DNA. It uses labeling and fragmentation coupled to gel electrophoresis to identify areas of the genome that have been bound by proteins.[27]
- MNase-seq (Micrococcal Nuclease sequencing) uses the micrococcal nuclease enzyme to identify nucleosome positioning throughout the genome.[28][29]
- Chromosome conformation capture determines the spatial organization of chromatin in the nucleus, by inferring genomic locations that physically interact.
- MACC profiling (Micrococcal nuclease ACCessibility profiling) uses titration series of chromatin digests with micrococcal nuclease to identify chromatin accessibility as well as to map nucleosomes and non-histone DNA-binding proteins in both open and closed regions of the genome.[30]
Chromatin and knots
It has been a puzzle how decondensed interphase chromosomes remain essentially unknotted. The natural expectation is that in the presence of type II DNA topoisomerases that permit passages of double-stranded DNA regions through each other, all chromosomes should reach the state of topological equilibrium. The topological equilibrium in highly crowded interphase chromosomes forming chromosome territories would result in formation of highly knotted chromatin fibres. However, Chromosome Conformation Capture (3C) methods revealed that the decay of contacts with the genomic distance in interphase chromosomes is practically the same as in the crumpled globule state that is formed when long polymers condense without formation of any knots. To remove knots from highly crowded chromatin, one would need an active process that should not only provide the energy to move the system from the state of topological equilibrium but also guide topoisomerase-mediated passages in such a way that knots would be efficiently unknotted instead of making the knots even more complex. It has been shown that the process of chromatin-loop extrusion is ideally suited to actively unknot chromatin fibres in interphase chromosomes.[31]
Chromatin: alternative definitions
The term, introduced by Walther Flemming, has multiple meanings:
- Simple and concise definition: Chromatin is a macromolecular complex of a DNA macromolecule and protein macromolecules (and RNA). The proteins package and arrange the DNA and control its functions within the cell nucleus.
- A biochemists’ operational definition: Chromatin is the DNA/protein/RNA complex extracted from eukaryotic lysed interphase nuclei. Just which of the multitudinous substances present in a nucleus will constitute a part of the extracted material partly depends on the technique each researcher uses. Furthermore, the composition and properties of chromatin vary from one cell type to the another, during development of a specific cell type, and at different stages in the cell cycle.
- The DNA + histone = chromatin definition: The DNA double helix in the cell nucleus is packaged by special proteins termed histones. The formed protein/DNA complex is called chromatin. The basic structural unit of chromatin is the nucleosome.
The first definition allows for "chromatins" to be defined in other domains of life like bacteria and archaea, using any DNA-binding proteins that condenses the molecule. These proteins are usually referred to nucleoid-associated proteins (NAPs); examples include AsnC/LrpC with HU. In addition, some archaea do produce nucleosomes from proteins homologous to eukaryotic histones.[32]
Nobel Prizes
The following scientists were recognized for their contributions to chromatin research with Nobel Prizes:
| Year | Who | Award |
|---|---|---|
| 1910 | Albrecht Kossel (University of Heidelberg) | Nobel Prize in Physiology or Medicine for his discovery of the five nuclear bases: adenine, cytosine, guanine, thymine, and uracil. |
| 1933 | Thomas Hunt Morgan (California Institute of Technology) | Nobel Prize in Physiology or Medicine for his discoveries of the role played by the gene and chromosome in heredity, based on his studies of the white-eyed mutation in the fruit fly Drosophila.[33] |
| 1962 | Francis Crick, James Watson and Maurice Wilkins (MRC Laboratory of Molecular Biology, Harvard University and London University respectively) | Nobel Prize in Physiology or Medicine for their discoveries of the double helix structure of DNA and its significance for information transfer in living material. |
| 1982 | Aaron Klug (MRC Laboratory of Molecular Biology) | Nobel Prize in Chemistry "for his development of crystallographic electron microscopy and his structural elucidation of biologically important nucleic acid-protein complexes" |
| 1993 | Richard J. Roberts and Phillip A. Sharp | Nobel Prize in Physiology "for their independent discoveries of split genes," in which DNA sections called exons express proteins, and are interrupted by DNA sections called introns, which do not express proteins. |
| 2006 | Roger Kornberg (Stanford University) | Nobel Prize in Chemistry for his discovery of the mechanism by which DNA is transcribed into messenger RNA. |
See also
Notes
References
- Monday, Tanmoy (July 2010). "Characterization of the RNA content of chromatin". Genome Res. 20 (7): 899–907. doi:10.1101/gr.103473.109. PMC 2892091. PMID 20404130.
- Hansen, Jeffrey (March 2012). "Human mitotic chromosome structure: what happened to the 30-nm fibre?". The EMBO Journal. 31 (7): 1621–1623. doi:10.1038/emboj.2012.66. PMC 3321215. PMID 22415369.
- Dame, R.T. (May 2005). "The role of nucleoid-associated proteins in the organization and compaction of bacterial chromatin". Molecular Microbiology. 56 (4): 858–870. doi:10.1111/j.1365-2958.2005.04598.x. PMID 15853876. S2CID 26965112.
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES (April 2006). "A bivalent chromatin structure marks key developmental genes in embryonic stem cells". Cell. 125 (2): 315–26. doi:10.1016/j.cell.2006.02.041. ISSN 0092-8674. PMID 16630819. S2CID 9993008.
- Portoso M, Cavalli G (2008). "The Role of RNAi and Noncoding RNAs in Polycomb Mediated Control of Gene Expression and Genomic Programming". RNA and the Regulation of Gene Expression: A Hidden Layer of Complexity. Caister Academic Press. ISBN 978-1-904455-25-7.
- Annunziato, Anthony T. "DNA Packaging: Nucleosomes and Chromatin". Scitable. Nature Education. Retrieved 2015-10-29.
- Robinson DJ; Fairall L; Huynh VA; Rhodes D. (April 2006). "EM measurements define the dimensions of the "30-nm" chromatin fiber: Evidence for a compact, interdigitated structure". Proceedings of the National Academy of Sciences of the United States of America. 103 (17): 6506–11. Bibcode:2006PNAS..103.6506R. doi:10.1073/pnas.0601212103. PMC 1436021. PMID 16617109.
-
Wong H, Victor JM, Mozziconacci J (September 2007). Chen P (ed.). "An All-Atom Model of the Chromatin Fiber Containing Linker Histones Reveals a Versatile Structure Tuned by the Nucleosomal Repeat Length". PLoS ONE. 2 (9): e877. Bibcode:2007PLoSO...2..877W. doi:10.1371/journal.pone.0000877. PMC 1963316. PMID 17849006.

- Nicodemi M, Pombo A (June 2014). "Models of chromosome structure" (PDF). Curr. Opin. Cell Biol. 28: 90–5. doi:10.1016/j.ceb.2014.04.004. PMID 24804566.
- Nicodemi M, Panning B, Prisco A (May 2008). "A thermodynamic switch for chromosome colocalization". Genetics. 179 (1): 717–21. arXiv:0809.4788. doi:10.1534/genetics.107.083154. PMC 2390650. PMID 18493085.
- Bohn M, Heermann DW (2010). "Diffusion-driven looping provides a consistent framework for chromatin organization". PLOS ONE. 5 (8): e12218. Bibcode:2010PLoSO...512218B. doi:10.1371/journal.pone.0012218. PMC 2928267. PMID 20811620.
- Lodish, Harvey F. (2016). Molecular Cell Biology (8th ed.). New York: W. H. Freeman and Company. p. 339. ISBN 978-1-4641-8339-3.
- Poonperm, R; Takata, H; Hamano, T; Matsuda, A; Uchiyama, S; Hiraoka, Y; Fukui, K (1 July 2015). "Chromosome Scaffold is a Double-Stranded Assembly of Scaffold Proteins". Scientific Reports. 5: 11916. doi:10.1038/srep11916. PMC 4487240. PMID 26132639.
- Xing H, Vanderford NL, Sarge KD (November 2008). "The TBP-PP2A mitotic complex bookmarks genes by preventing condensin action". Nat. Cell Biol. 10 (11): 1318–23. doi:10.1038/ncb1790. PMC 2577711. PMID 18931662.
- Allfrey VG, Faulkner R, Mirsky AE (May 1964). "Acetylation and Methylation of Histones and Their Possible Role in the Regulation of RNA Synthesis". Proc. Natl. Acad. Sci. U.S.A. 51 (5): 786–94. Bibcode:1964PNAS...51..786A. doi:10.1073/pnas.51.5.786. PMC 300163. PMID 14172992.
- Kaochar S, Tu BP (November 2012). "Gatekeepers of chromatin: Small metabolites elicit big changes in gene expression". Trends Biochem. Sci. 37 (11): 477–83. doi:10.1016/j.tibs.2012.07.008. PMC 3482309. PMID 22944281.
- De Vries M, Ramos L, Housein Z, De Boer P (May 2012). "Chromatin remodelling initiation during human spermiogenesis". Biol Open. 1 (5): 446–57. doi:10.1242/bio.2012844. PMC 3507207. PMID 23213436.
- Murugesapillai D, McCauley MJ, Huo R, Nelson Holte MH, Stepanyants A, Maher LJ, Israeloff NE, Williams MC (August 2014). "DNA bridging and looping by HMO1 provides a mechanism for stabilizing nucleosome-free chromatin". Nucleic Acids Research. 42 (14): 8996–9004. doi:10.1093/nar/gku635. PMC 4132745. PMID 25063301.
- Murugesapillai D, McCauley MJ, Maher LJ, Williams MC (February 2017). "Single-molecule studies of high-mobility group B architectural DNA bending proteins". Biophysical Reviews. 9 (1): 17–40. doi:10.1007/s12551-016-0236-4. PMC 5331113. PMID 28303166.
- Liu B, Yip RK, Zhou Z (2012). "Chromatin remodeling, DNA damage repair and aging". Curr. Genomics. 13 (7): 533–47. doi:10.2174/138920212803251373. PMC 3468886. PMID 23633913.
- Sellou H, Lebeaupin T, Chapuis C, Smith R, Hegele A, Singh HR, Kozlowski M, Bultmann S, Ladurner AG, Timinszky G, Huet S (2016). "The poly(ADP-ribose)-dependent chromatin remodeler Alc1 induces local chromatin relaxation upon DNA damage". Mol. Biol. Cell. 27 (24): 3791–3799. doi:10.1091/mbc.E16-05-0269. PMC 5170603. PMID 27733626.
- Haince JF, McDonald D, Rodrigue A, Déry U, Masson JY, Hendzel MJ, Poirier GG (2008). "PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites". J. Biol. Chem. 283 (2): 1197–208. doi:10.1074/jbc.M706734200. PMID 18025084.
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM (1998). "DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139". J. Biol. Chem. 273 (10): 5858–68. doi:10.1074/jbc.273.10.5858. PMID 9488723.
- Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J (2007). "RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins". Cell. 131 (5): 887–900. doi:10.1016/j.cell.2007.09.040. PMID 18001824. S2CID 14232192.
- Luijsterburg MS, Acs K, Ackermann L, Wiegant WW, Bekker-Jensen S, Larsen DH, Khanna KK, van Attikum H, Mailand N, Dantuma NP (2012). "A new non-catalytic role for ubiquitin ligase RNF8 in unfolding higher-order chromatin structure". EMBO J. 31 (11): 2511–27. doi:10.1038/emboj.2012.104. PMC 3365417. PMID 22531782.
- Giresi, Paul G.; Kim, Jonghwan; McDaniell, Ryan M.; Iyer, Vishwanath R.; Lieb, Jason D. (2007-06-01). "FAIRE (Formaldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin". Genome Research. 17 (6): 877–885. doi:10.1101/gr.5533506. ISSN 1088-9051. PMC 1891346. PMID 17179217.
- Galas, D. J.; Schmitz, A. (1978-09-01). "DNAse footprinting: a simple method for the detection of protein-DNA binding specificity". Nucleic Acids Research. 5 (9): 3157–3170. doi:10.1093/nar/5.9.3157. ISSN 0305-1048. PMC 342238. PMID 212715.
- Cui, Kairong; Zhao, Keji (2012-01-01). Genome-wide approaches to determining nucleosome occupancy in metazoans using MNase-Seq. Methods in Molecular Biology. 833. pp. 413–419. doi:10.1007/978-1-61779-477-3_24. ISBN 978-1-61779-476-6. ISSN 1940-6029. PMC 3541821. PMID 22183607.
- Buenrostro, Jason D.; Giresi, Paul G.; Zaba, Lisa C.; Chang, Howard Y.; Greenleaf, William J. (2013-12-01). "Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position". Nature Methods. 10 (12): 1213–1218. doi:10.1038/nmeth.2688. ISSN 1548-7105. PMC 3959825. PMID 24097267.
- Mieczkowski J, Cook A, Bowman SK, Mueller B, Alver BH, Kundu S, Deaton AM, Urban JA, Larschan E, Park PJ, Kingston RE, Tolstorukov MY (2016-05-06). "MNase titration reveals differences between nucleosome occupancy and chromatin accessibility". Nature Communications. 7: 11485. Bibcode:2016NatCo...711485M. doi:10.1038/ncomms11485. PMC 4859066. PMID 27151365.
- Racko D, Benedetti F, Goundaroulis D, Stasiak A (2018). "Chromatin Loop Extrusion and Chromatin Unknotting". Polymers. 10 (10): 1126–1137. doi:10.3390/polym10101126. PMC 6403842. PMID 30961051.
- Luijsterburg, Martijn S.; White, Malcolm F.; van Driel, Roel; Dame, Remus Th. (8 January 2009). "The Major Architects of Chromatin: Architectural Proteins in Bacteria, Archaea and Eukaryotes". Critical Reviews in Biochemistry and Molecular Biology. 43 (6): 393–418. doi:10.1080/10409230802528488. PMID 19037758. S2CID 85874882.
- "Thomas Hunt Morgan and His Legacy". Nobelprize.org. 7 Sep 2012
Additional sources
- Cooper, Geoffrey M. 2000. The Cell, 2nd edition, A Molecular Approach. Chapter 4.2 Chromosomes and Chromatin.
- Corces, V. G. (1995). "Chromatin insulators. Keeping enhancers under control". Nature. 376 (6540): 462–463. Bibcode:1995Natur.376..462C. doi:10.1038/376462a0. PMID 7637775. S2CID 26494996.
- Cremer, T. 1985. Von der Zellenlehre zur Chromosomentheorie: Naturwissenschaftliche Erkenntnis und Theorienwechsel in der frühen Zell- und Vererbungsforschung, Veröffentlichungen aus der Forschungsstelle für Theoretische Pathologie der Heidelberger Akademie der Wissenschaften. Springer-Vlg., Berlin, Heidelberg.
- Elgin, S. C. R. (ed.). 1995. Chromatin Structure and Gene Expression, vol. 9. IRL Press, Oxford, New York, Tokyo.
- Gerasimova, T. I.; Corces, V. G. (1996). "Boundary and insulator elements in chromosomes". Curr. Opin. Genet. Dev. 6 (2): 185–192. doi:10.1016/s0959-437x(96)80049-9. PMID 8722175.
- Gerasimova, T. I.; Corces, V. G. (1998). "Polycomb and Trithorax group proteins mediate the function of a chromatin insulator". Cell. 92 (4): 511–521. doi:10.1016/s0092-8674(00)80944-7. PMID 9491892. S2CID 8192263.
- Gerasimova, T. I.; Corces, V. G. (2001). "CHROMATIN INSULATORS AND BOUNDARIES: Effects on Transcription and Nuclear Organization". Annu Rev Genet. 35: 193–208. doi:10.1146/annurev.genet.35.102401.090349. PMID 11700282. S2CID 22738830.
- Gerasimova, T. I.; Byrd, K.; Corces, V. G. (2000). "A chromatin insulator determines the nuclear localization of DNA [In Process Citation]". Mol Cell. 6 (5): 1025–35. doi:10.1016/s1097-2765(00)00101-5. PMID 11106742.
- Ha, S. C.; Lowenhaupt, K.; Rich, A.; Kim, Y. G.; Kim, K. K. (2005). "Crystal structure of a junction between B-DNA and Z-DNA reveals two extruded bases". Nature. 437 (7062): 1183–6. Bibcode:2005Natur.437.1183H. doi:10.1038/nature04088. PMID 16237447. S2CID 2539819.
- Pollard, T., and W. Earnshaw. 2002. Cell Biology. Saunders.
- Saumweber, H. 1987. Arrangement of Chromosomes in Interphase Cell Nuclei, p. 223-234. In W. Hennig (ed.), Structure and Function of Eucaryotic Chromosomes, vol. 14. Springer-Verlag, Berlin, Heidelberg.
- Sinden, R. R. (2005). "Molecular biology: DNA twists and flips". Nature. 437 (7062): 1097–8. doi:10.1038/4371097a. PMID 16237426. S2CID 4409092.
- Van Holde KE. 1989. Chromatin. New York: Springer-Verlag. ISBN 0-387-96694-3.
- Van Holde, K., J. Zlatanova, G. Arents, and E. Moudrianakis. 1995. Elements of chromatin structure: histones, nucleosomes, and fibres, p. 1-26. In S. C. R. Elgin (ed.), Chromatin structure and gene expression. IRL Press at Oxford University Press, Oxford.
External links
- Chromatin, Histones & Cathepsin; PMAP The Proteolysis Map-animation
- [ Recent chromatin publications and news]
- Protocol for in vitro Chromatin Assembly
- ENCODE threads Explorer Chromatin patterns at transcription factor binding sites. Nature (journal)