LIM domain
LIM domains are protein structural domains, composed of two contiguous zinc finger domains, separated by a two-amino acid residue hydrophobic linker.[1] The structure of these domains varies depending on type such as cysteine-rich LIM (LIN-11,[2] Isl-1[2] and MEC-3[2]) domains contain certain tetrahedral coordinations at S3N and S4.[2]
| LIM domain | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
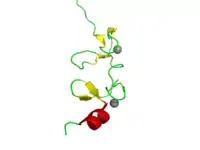 Structure of the 4th LIM domain of Pinch protein. Zinc atoms are shown in grey | |||||||||||
| Identifiers | |||||||||||
| Symbol | LIM | ||||||||||
| Pfam | PF00412 | ||||||||||
| InterPro | IPR001781 | ||||||||||
| PROSITE | PDOC50178 | ||||||||||
| SCOP2 | 1ctl / SCOPe / SUPFAM | ||||||||||
| CDD | cd08368 | ||||||||||
| |||||||||||
Discovery
LIM domains are named after their initial discovery in the three proteins that have the following functions;[3][2]
Roles
LIM-domain containing proteins have been shown to play roles in cytoskeletal organization, organ development, regulation plant cell development, cell lineage specification, and regulation of gene transcription.[4] The dysfunctional roles of LIM domains are oncogenesis, embryonic lethality, and muscle detachment.[4] LIM-domains mediate protein–protein interactions that are critical to cellular processes. For example, direct force-activated F-actin binding by LIM Domains is a mechanosensing mechanism by which cytoskeletal tension can govern nuclear localization.[5] This typically involves acting as a conserved scaffold for recognizing target proteins.[4]
Sequencing and Protein Interaction
LIM domains have highly divergent sequences, apart from certain key residues. The sequence divergence allow a great many different binding sites to be grafted onto the same basic domain. The conserved residues are those involved in zinc binding or the hydrophobic core of the protein. The two zinc ions will primarily serve for the structure of the domain as seen in CRP1 and CRIP.[4] Also, the domains have specific affinity for zinc.[4] The sequence signature of LIM domains is as follows:
[C]-[X]2–4-[C]-[X]13–19-[W]-[H]-[X]2–4-[C]-[F]-[LVI]-[C]-[X]2–4-[C]-[X]13–20-C-[X]2–4-[C]
LIM domains frequently occur in multiples, as seen in proteins such as TES, LMO4, and can also be attached to other domains in order to confer a binding or targeting function upon them, such as LIM-kinase.
Classification
The LIM superclass of genes have been classified into 14 classes: ABLIM, CRP, ENIGMA, EPLIN, LASP, LHX, LMO, LIMK, LMO7, MICAL, PXN, PINCH, TES, and ZYX. Six of these classes (i.e., ABLIM, MICAL, ENIGMA, ZYX, LHX, LM07) originated in the stem lineage of animals, and this expansion is thought to have made a major contribution to the origin of animal multicellularity.[6]
Asides lineage of animals, there are an entire class of plan LIM genes that were classified into four different classes: WLIM1, WLIM2, PLIM1, PLIM2, and FLIM (XLIM).[7] These are sorted into 4 different subfamilies: αLIM1, βLIM1, γLIM2, and δLIM2.[7] The αLIM1 subclades include PLIM1, WLIM1, and FLIM (XLIM).[7] βLIM1 is a new subfamily, so no current distinguishable subclades.[7] γLIM2 subclades contain WLIM2 and PLIM2.[7] The final subfamily δLIM2 contains WLIM2, and PLIM2.[7]
LIM domains are also found in various bacterial lineages where they are typically fused to a metallopeptidase domain. Some versions show fusions to an inactive P-loop NTPase at their N-terminus and a single transmembrane helix. These domain fusions suggest that the prokaryotic LIM domains are likely to regulate protein processing at the cell membrane. The domain architectural syntax is remarkably parallel to those of the prokaryotic versions of the B-box zinc finger and the AN1 zinc finger domains.[8]
LIM domain containing proteins serve many specific functions in cells such as adherens junction, cytoarchitecture, specification of cell polarity, nuclear-cytoplasmic shuttling, and protein trafficking.[3] These domains can be found in eukaryoes, plants, animal, fungi, and mycetozoa.[4] It was classified as A, B, C, and D.[4] These classifications are further sorted into three groups.
Group 1
This group contains LIM domain classes A and B.[4] They are typically fused to other functional domains such as kinases.[4] The subclasses for these domains are LIM-homeodomain transcription factors, LMO proteins, and LIM kinases.[4]
LIM-homeodomain transcription factors
They have multifunctionality primarily focusing on development of the nervous system, activation of transcription, and cell fate specification during development.[3][4] The nervous system relies on the LIM domain type for differentiation of neurotransmitter biosynthetic pathways.[3]
LMO proteins
These proteins focus on overall development of multiple cell types as well as oncogenesis and transcriptional regulation.[3] Oncogenesis was found to occur due to the expression of LMO 1 and LMO 2 in T-cell leukemia patients.[3][4]
LIM kinases
The purpose of these proteins is the establishment and regulation of the cytoskeleton.[4] The regulation of the cytoskeleton by these kinases is through phosphorylation of cofilin, which allows for the accumulation of actin filaments.[4] Notably, they have been found to be responsible for regulation of cell motility and morphogenesis.[3]
Group 2
This group contains LIM domain class C, which are localized typically in the cytoplasm.[3][4] These domains are internally duplicated with two copies per a protein.[4] Also, they are more similar to each than classes A and B.[4]
Cysteine-rich proteins
There are three different cysteine rich proteins.[4] The purpose of these proteins is their role in myogenesis and muscle structure.[4] Although, it was found that structural role is played in multiple types of cells.[4] Each of the CRP proteins are activated throughout myogenesis.[4] CRP 3 plays a role in development of myoblasts, while CRP 1 is active in fibro blast cells.[4] CRP 1 has more roles involved with actin filaments and z lines of myofibers.[3]
Group 3
This group contains only class D, which are typically localized in the cytoplasm.[4][3] These LIM proteins contain 1 to 5 domains.[4] These domains can have additional functional domains or motifs.[4] This group is limited to three different adaptor proteins: zyxin, paxillin, and PINCH.[4] They each respectively have different number of LIM domains with 3, 4, and 5.[4] These are considered adaptor proteins related to adhesion plaques that regulate cell shape and spreading through distinct LIM-mediated protein-protein interactions.[4]
Protein-protein interactions
LIM-HD & LMO
These proteins are formed through interaction of LIM domain binding families that are bound by LIM1.[4] LIM-Ldb interacts to form different heterodimers of LIM-HD.[4] THis will typically form a LIM-LID region that interacts with LIM proteins.[4] LIM-HD is known to determine distinct identities for motor neurons during development.[4] It has been found to bind LMO1, LMO2, Lhx1, Isl1, and Mec3.[4] LMO2 is found to be localized in the nucleus, which is involved in erythroid development especially in the fetal liver.[3][2]
Zyxin
This protein is localized between the cytoplasm and nucleus through shuttling.[3] It focuses on moving between cell adhesion sites and nucleus.[3] The zinc fingers of the LIM domain will act independently.[4] Zyxin has a variety of bind partners such as CRP, α-actinin, proto-oncogene Vav, p130, and members of Ena/VASP family of proteins.[4] The known interactions of zyxin are between Ena/VASP and CRP1.[4] LIM1 is responsible for recognition of CRP1, but cooperate with LIM2 for binding to zyxin.[4] The Ena/VASP will bind profilin that is known to act as a actin-polymerizing protein.[4] The zyxin-VASP complex will initiate actin-polymerization for the cytoskeletal strucutre.[4][2]
Paxillin
This protein is localized in the cytoplasm at focal adhesion sites.[3] It functions as a central protein for fatty acids and development of cystoskeletal structure.[4][2] In fatty acids, they act as scaffolds for many binding partners.[4] The LIM domain at the c-terminal bind protein tyrosine phosphatase-PEST.[4] PTP-PEST binds at c-termini LIM 3 and 4 to disassemble fatty acids that will lead to the modulation of the fatty acid targeting regions.[4] The extent of binding will depend on LIM 2 and 4.[4] This will occur upon dephosphorylation of p130 and paxillin.[4]
ENIGMA
This protein is localized in the cytoplasm, which serves in signaling and protein trafficking.[3][2] The structure of this protein contains three LIM domains at the c-terminal.[4] It will bind insulin receptor internalization motif (InsRF) at LIM domain 3.[4] LIM domain 2 binds Ret receptor tyrosine kinase.[4]
PINCH
This protein is localized in the cytoplasm and nucleus.[3] It is responsible for effecting specific muscle adherens junctions and mechanosensory functions of touch receptor neurons.[3] The protein sequence in the LIM domains are linked with very short interdomain peptides and c-terminal extension with high amounts of positive charges.[4] The protein has multiple functions even presenting in senescent enrythrocyte antigens.[4] It can bind to ankyrin repeat domains of integrin-linked kinase.[4] Also, LIM domain 4 of PINCH can bind to Nck2 protein to act as a adaptor.[4]
References
- Kadrmas JL, Beckerle MC (November 2004). "The LIM domain: from the cytoskeleton to the nucleus". Nature Reviews. Molecular Cell Biology. 5 (11): 920–31. doi:10.1038/nrm1499. PMID 15520811. S2CID 6030950.
- Gill GN (December 1995). "The enigma of LIM domains". Structure. 3 (12): 1285–9. doi:10.1016/S0969-2126(01)00265-9. PMID 8747454.
- Bach I (March 2000). "The LIM domain: regulation by association". Mechanisms of Development. 91 (1–2): 5–17. doi:10.1016/S0925-4773(99)00314-7. PMID 10704826. S2CID 16093470.
- Iuchi S, Kuldell N (6 March 2007). Zinc Finger Proteins: From Atomic Contact to Cellular Function. Springer Science & Business Media. ISBN 978-0-387-27421-8.
- Sun, X., Phua, D. Y., Axiotakis, L., Smith, M. A., Blankman, E., Gong, R., ... & Alushin, G. M. (2020). Mechanosensing through direct binding of tensed F-actin by LIM domains. Developmental Cell. doi:10.1016/j.devcel.2020.09.022
- Koch BJ, Ryan JF, Baxevanis AD (March 2012). "The diversification of the LIM superclass at the base of the metazoa increased subcellular complexity and promoted multicellular specialization". PLOS ONE. 7 (3): e33261. Bibcode:2012PLoSO...733261K. doi:10.1371/journal.pone.0033261. PMC 3305314. PMID 22438907.
- Cheng X, Li G, Muhammad A, Zhang J, Jiang T, Jin Q, et al. (February 2019). "Molecular identification, phylogenomic characterization and expression patterns analysis of the LIM (LIN-11, Isl1 and MEC-3 domains) gene family in pear (Pyrus bretschneideri) reveal its potential role in lignin metabolism". Gene. 686: 237–249. doi:10.1016/j.gene.2018.11.064. PMID 30468911.
- Burroughs AM, Iyer LM, Aravind L (July 2011). "Functional diversification of the RING finger and other binuclear treble clef domains in prokaryotes and the early evolution of the ubiquitin system". Molecular BioSystems. 7 (7): 2261–77. doi:10.1039/C1MB05061C. PMC 5938088. PMID 21547297.
