MRAS
Ras-related protein M-Ras, also known as muscle RAS oncogene homolog and R-Ras3, is a protein that in humans is encoded by the MRAS gene on chromosome 3.[5][6][7] It is ubiquitously expressed in many tissues and cell types.[8] This protein functions as a signal transducer for a wide variety of signaling pathways, including those promoting neural and bone formation as well as tumor growth.[9][10][11][12] The MRAS gene also contains one of 27 SNPs associated with increased risk of coronary artery disease.[13]
Structure
Gene
The MRAS gene resides on chromosome 3 at the band 3q22.3 and includes 10 exons.[7] This gene produces 2 isoforms through alternative splicing.[14]
Protein
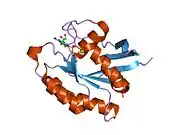
M-Ras is a member of the small GTPase superfamily under the Ras family, which also includes Rap1, Rap2, R-Ras, and R-Ras2 (TC21).[14] This protein spans a length of 209 residues. Its N-terminal amino acid sequence shares 60-75% identity with that in the Ras protein while its effector region is identical with that in Ras. M-Ras shares a similar structure with H-Ras and Rap2A with the exception of its switch 1 conformation when bound to guanosine 5'-(beta,gamma-imido)triphosphate (Gpp(NH)p). Of the two states M-Ras can switch between, M-Ras is predominantly found in its state 1 conformation, which does not bind Ras effectors.[15]
Function
The MRAS gene is expressed specifically in brain, heart, myoblasts, myotubes, fibroblasts, skeletal muscles, and uterus, suggesting a specific role of M-Ras in these tissue and cells.[16][17] M-Ras is involved in many biological processes by activating a wide variety of proteins. For instance, it is activated by Ras guanine nucleotide exchange factors and can bind/activate some Ras protein effectors.[18] M-Ras also weakly stimulates the mitogen-activated protein kinase (MAPK) activity and ERK2 activity, but modestly stimulates trans-activation from different nuclear response elements which bind transcription factors, such as SRF, ETS/TCF, Jun/Fos, and NF- kB/Rel.[17][19] M-Ras has been found to induce Akt kinase activity in the PI3-K pathway, and it may play a role in cell survival of neural-derived cells.[20] Moreover, M-Ras plays a crucial role in the downregulation of OCT4 and NANOG protein levels upon differentiation and has been demonstrated to modulate cell fate at early steps of development during neurogenesis.[21] M-Ras, induced and activated by BMP-2 signaling, also participates in the osteoblastic determination, differentiation, and transdifferentiation under p38 MAPK and JNK regulation.[22] M-Ras is involved in TNF-alpha-stimulated and Rap1-mediated LFA-1 activation in splenocytes.[23] More generally, cells transfected with M-Ras exhibit dendritic appearances with microspikes, suggesting that M-Ras may participate in reorganization of the actin cytoskeleton.[16] In addition, it is reported that M-Ras forms a complex with SCRIB and SHOC2, a polarity protein with tumor suppressor properties, and may play a key role in tumorigenic growth.[24]
Clinical significance
In humans, other members of the Ras subfamilies carry mutations in human cancers.[25] Furthermore, the Ras proteins are not only involved in tumorigenesis but also in many developmental disorders.[25] For instance, the Ras-related proteins appear to be overexpressed in human carcinomas of the oral cavity, esophagus, stomach, skin, and breast, as well as in lymphomas.[26][27][28][29] More currently, Ras family members such as R-RAS, R-RAS2 and also R-RAS3 have also been implicated as main factors in triggering neural transformation, with R-RAS2 as the most significant element.[30]
Clinical marker
A multi-locus genetic risk score study based on a combination of 27 loci, including the MRAS gene, identified individuals at increased risk for both incidence and recurrent coronary artery disease events, as well as an enhanced clinical benefit from statin therapy. The study was based on a community cohort study (the Malmo Diet and Cancer study) and four additional randomized controlled trials of primary prevention cohorts (JUPITER and ASCOT) and secondary prevention cohorts (CARE and PROVE IT-TIMI 22).[31]
References
- GRCh38: Ensembl release 89: ENSG00000158186 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000032470 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Kimmelman A, Tolkacheva T, Lorenzi MV, Osada M, Chan AM (November 1997). "Identification and characterization of R-ras3: a novel member of the RAS gene family with a non-ubiquitous pattern of tissue distribution". Oncogene. 15 (22): 2675–85. doi:10.1038/sj.onc.1201674. PMID 9400994.
- Quilliam LA, Castro AF, Rogers-Graham KS, Martin CB, Der CJ, Bi C (August 1999). "M-Ras/R-Ras3, a transforming ras protein regulated by Sos1, GRF1, and p120 Ras GTPase-activating protein, interacts with the putative Ras effector AF6". The Journal of Biological Chemistry. 274 (34): 23850–7. doi:10.1074/jbc.274.34.23850. PMID 10446149.
- "Entrez Gene: MRAS muscle RAS oncogene homolog".
- "BioGPS - your Gene Portal System". biogps.org. Retrieved 2016-10-10.
- Kimmelman AC, Osada M, Chan AM (April 2000). "R-Ras3, a brain-specific Ras-related protein, activates Akt and promotes cell survival in PC12 cells". Oncogene. 19 (16): 2014–22. doi:10.1038/sj.onc.1203530. PMID 10803462.
- Mathieu ME, Faucheux C, Saucourt C, Soulet F, Gauthereau X, Fédou S, Trouillas M, Thézé N, Thiébaud P, Boeuf H (August 2013). "MRAS GTPase is a novel stemness marker that impacts mouse embryonic stem cell plasticity and Xenopus embryonic cell fate". Development. 140 (16): 3311–22. doi:10.1242/dev.091082. PMID 23863483.
- Watanabe-Takano H, Takano K, Keduka E, Endo T (February 2010). "M-Ras is activated by bone morphogenetic protein-2 and participates in osteoblastic determination, differentiation, and transdifferentiation". Experimental Cell Research. 316 (3): 477–90. doi:10.1016/j.yexcr.2009.09.028. PMID 19800879.
- Young LC, Hartig N, Muñoz-Alegre M, Oses-Prieto JA, Durdu S, Bender S, Vijayakumar V, Vietri Rudan M, Gewinner C, Henderson S, Jathoul AP, Ghatrora R, Lythgoe MF, Burlingame AL, Rodriguez-Viciana P (December 2013). "An MRAS, SHOC2, and SCRIB complex coordinates ERK pathway activation with polarity and tumorigenic growth". Molecular Cell. 52 (5): 679–92. doi:10.1016/j.molcel.2013.10.004. PMID 24211266.
- Mega JL, Stitziel NO, Smith JG, Chasman DI, Caulfield MJ, Devlin JJ, Nordio F, Hyde CL, Cannon CP, Sacks FM, Poulter NR, Sever PS, Ridker PM, Braunwald E, Melander O, Kathiresan S, Sabatine MS (June 2015). "Genetic risk, coronary heart disease events, and the clinical benefit of statin therapy: an analysis of primary and secondary prevention trials". Lancet. 385 (9984): 2264–71. doi:10.1016/S0140-6736(14)61730-X. PMC 4608367. PMID 25748612.
- "MRAS - Ras-related protein M-Ras precursor - Homo sapiens (Human) - MRAS gene & protein". www.uniprot.org. Retrieved 2016-10-10.
- Ye M, Shima F, Muraoka S, Liao J, Okamoto H, Yamamoto M, Tamura A, Yagi N, Ueki T, Kataoka T (September 2005). "Crystal structure of M-Ras reveals a GTP-bound "off" state conformation of Ras family small GTPases". The Journal of Biological Chemistry. 280 (35): 31267–75. doi:10.1074/jbc.M505503200. PMID 15994326.
- Matsumoto K, Asano T, Endo T (November 1997). "Novel small GTPase M-Ras participates in reorganization of actin cytoskeleton". Oncogene. 15 (20): 2409–17. doi:10.1038/sj.onc.1201416. PMID 9395237.
- Kimmelman A, Tolkacheva T, Lorenzi MV, Osada M, Chan AM (November 1997). "Identification and characterization of R-ras3: a novel member of the RAS gene family with a non-ubiquitous pattern of tissue distribution". Oncogene. 15 (22): 2675–85. doi:10.1038/sj.onc.1201674. PMID 9400994.
- Rebhun JF, Castro AF, Quilliam LA (November 2000). "Identification of guanine nucleotide exchange factors (GEFs) for the Rap1 GTPase. Regulation of MR-GEF by M-Ras-GTP interaction". The Journal of Biological Chemistry. 275 (45): 34901–8. doi:10.1074/jbc.M005327200. PMID 10934204.
- Quilliam LA, Castro AF, Rogers-Graham KS, Martin CB, Der CJ, Bi C (August 1999). "M-Ras/R-Ras3, a transforming ras protein regulated by Sos1, GRF1, and p120 Ras GTPase-activating protein, interacts with the putative Ras effector AF6". The Journal of Biological Chemistry. 274 (34): 23850–7. doi:10.1074/jbc.274.34.23850. PMID 10446149.
- Kimmelman AC, Osada M, Chan AM (April 2000). "R-Ras3, a brain-specific Ras-related protein, activates Akt and promotes cell survival in PC12 cells". Oncogene. 19 (16): 2014–22. doi:10.1038/sj.onc.1203530. PMID 10803462.
- Mathieu ME, Faucheux C, Saucourt C, Soulet F, Gauthereau X, Fédou S, Trouillas M, Thézé N, Thiébaud P, Boeuf H (August 2013). "MRAS GTPase is a novel stemness marker that impacts mouse embryonic stem cell plasticity and Xenopus embryonic cell fate". Development. 140 (16): 3311–22. doi:10.1242/dev.091082. PMID 23863483.
- Watanabe-Takano H, Takano K, Keduka E, Endo T (February 2010). "M-Ras is activated by bone morphogenetic protein-2 and participates in osteoblastic determination, differentiation, and transdifferentiation". Experimental Cell Research. 316 (3): 477–90. doi:10.1016/j.yexcr.2009.09.028. PMID 19800879.
- Yoshikawa Y, Satoh T, Tamura T, Wei P, Bilasy SE, Edamatsu H, Aiba A, Katagiri K, Kinashi T, Nakao K, Kataoka T (August 2007). "The M-Ras-RA-GEF-2-Rap1 pathway mediates tumor necrosis factor-alpha dependent regulation of integrin activation in splenocytes". Molecular Biology of the Cell. 18 (8): 2949–59. doi:10.1091/mbc.E07-03-0250. PMC 1949361. PMID 17538012.
- Young LC, Hartig N, Muñoz-Alegre M, Oses-Prieto JA, Durdu S, Bender S, Vijayakumar V, Vietri Rudan M, Gewinner C, Henderson S, Jathoul AP, Ghatrora R, Lythgoe MF, Burlingame AL, Rodriguez-Viciana P (December 2013). "An MRAS, SHOC2, and SCRIB complex coordinates ERK pathway activation with polarity and tumorigenic growth". Molecular Cell. 52 (5): 679–92. doi:10.1016/j.molcel.2013.10.004. PMID 24211266.
- Karnoub AE, Weinberg RA (July 2008). "Ras oncogenes: split personalities". Nature Reviews Molecular Cell Biology. 9 (7): 517–31. doi:10.1038/nrm2438. PMC 3915522. PMID 18568040.
- Graham SM, Oldham SM, Martin CB, Drugan JK, Zohn IE, Campbell S, Der CJ (March 1999). "TC21 and Ras share indistinguishable transforming and differentiating activities". Oncogene. 18 (12): 2107–16. doi:10.1038/sj.onc.1202517. PMID 10321735.
- Cox AD, Brtva TR, Lowe DG, Der CJ (November 1994). "R-Ras induces malignant, but not morphologic, transformation of NIH3T3 cells". Oncogene. 9 (11): 3281–8. PMID 7936652.
- Chan AM, Miki T, Meyers KA, Aaronson SA (August 1994). "A human oncogene of the RAS superfamily unmasked by expression cDNA cloning". Proceedings of the National Academy of Sciences of the United States of America. 91 (16): 7558–62. doi:10.1073/pnas.91.16.7558. PMC 44441. PMID 8052619.
- Huang Y, Saez R, Chao L, Santos E, Aaronson SA, Chan AM (October 1995). "A novel insertional mutation in the TC21 gene activates its transforming activity in a human leiomyosarcoma cell line". Oncogene. 11 (7): 1255–60. PMID 7478545.
- Gutierrez-Erlandsson S, Herrero-Vidal P, Fernandez-Alfara M, Hernandez-Garcia S, Gonzalo-Flores S, Mudarra-Rubio A, Fresno M, Cubelos B (2013-01-01). "R-RAS2 overexpression in tumors of the human central nervous system". Molecular Cancer. 12 (1): 127. doi:10.1186/1476-4598-12-127. PMC 3900289. PMID 24148564.
- Mega JL, Stitziel NO, Smith JG, Chasman DI, Caulfield MJ, Devlin JJ, Nordio F, Hyde CL, Cannon CP, Sacks FM, Poulter NR, Sever PS, Ridker PM, Braunwald E, Melander O, Kathiresan S, Sabatine MS (June 2015). "Genetic risk, coronary heart disease events, and the clinical benefit of statin therapy: an analysis of primary and secondary prevention trials". Lancet. 385 (9984): 2264–71. doi:10.1016/S0140-6736(14)61730-X. PMC 4608367. PMID 25748612.
- Ortiz-Vega S, Khokhlatchev A, Nedwidek M, Zhang XF, Dammann R, Pfeifer GP, Avruch J (February 2002). "The putative tumor suppressor RASSF1A homodimerizes and heterodimerizes with the Ras-GTP binding protein Nore1". Oncogene. 21 (9): 1381–90. doi:10.1038/sj.onc.1205192. PMID 11857081.
- Ehrhardt GR, Leslie KB, Lee F, Wieler JS, Schrader JW (October 1999). "M-Ras, a widely expressed 29-kD homologue of p21 Ras: expression of a constitutively active mutant results in factor-independent growth of an interleukin-3-dependent cell line". Blood. 94 (7): 2433–44. doi:10.1182/blood.V94.7.2433.419k31_2433_2444. PMID 10498616.
Further reading
- Matsumoto K, Asano T, Endo T (November 1997). "Novel small GTPase M-Ras participates in reorganization of actin cytoskeleton". Oncogene. 15 (20): 2409–17. doi:10.1038/sj.onc.1201416. PMID 9395237.
- Louahed J, Grasso L, De Smet C, Van Roost E, Wildmann C, Nicolaides NC, Levitt RC, Renauld JC (September 1999). "Interleukin-9-induced expression of M-Ras/R-Ras3 oncogene in T-helper clones". Blood. 94 (5): 1701–10. doi:10.1182/blood.V94.5.1701. PMID 10477695.
- Ehrhardt GR, Leslie KB, Lee F, Wieler JS, Schrader JW (October 1999). "M-Ras, a widely expressed 29-kD homologue of p21 Ras: expression of a constitutively active mutant results in factor-independent growth of an interleukin-3-dependent cell line". Blood. 94 (7): 2433–44. doi:10.1182/blood.V94.7.2433.419k31_2433_2444. PMID 10498616.
- Kimmelman AC, Osada M, Chan AM (April 2000). "R-Ras3, a brain-specific Ras-related protein, activates Akt and promotes cell survival in PC12 cells". Oncogene. 19 (16): 2014–22. doi:10.1038/sj.onc.1203530. PMID 10803462.
- Rebhun JF, Castro AF, Quilliam LA (November 2000). "Identification of guanine nucleotide exchange factors (GEFs) for the Rap1 GTPase. Regulation of MR-GEF by M-Ras-GTP interaction". The Journal of Biological Chemistry. 275 (45): 34901–8. doi:10.1074/jbc.M005327200. PMID 10934204.
- Gao X, Satoh T, Liao Y, Song C, Hu CD, Kariya Ki K, Kataoka T (November 2001). "Identification and characterization of RA-GEF-2, a Rap guanine nucleotide exchange factor that serves as a downstream target of M-Ras". The Journal of Biological Chemistry. 276 (45): 42219–25. doi:10.1074/jbc.M105760200. PMID 11524421.
- Ortiz-Vega S, Khokhlatchev A, Nedwidek M, Zhang XF, Dammann R, Pfeifer GP, Avruch J (February 2002). "The putative tumor suppressor RASSF1A homodimerizes and heterodimerizes with the Ras-GTP binding protein Nore1". Oncogene. 21 (9): 1381–90. doi:10.1038/sj.onc.1205192. PMID 11857081.
- Kimmelman AC, Nuñez Rodriguez N, Chan AM (August 2002). "R-Ras3/M-Ras induces neuronal differentiation of PC12 cells through cell-type-specific activation of the mitogen-activated protein kinase cascade". Molecular and Cellular Biology. 22 (16): 5946–61. doi:10.1128/MCB.22.16.5946-5961.2002. PMC 133986. PMID 12138204.
- Mitin NY, Ramocki MB, Zullo AJ, Der CJ, Konieczny SF, Taparowsky EJ (May 2004). "Identification and characterization of rain, a novel Ras-interacting protein with a unique subcellular localization". The Journal of Biological Chemistry. 279 (21): 22353–61. doi:10.1074/jbc.M312867200. PMID 15031288.
- Roberts AE, Araki T, Swanson KD, Montgomery KT, Schiripo TA, Joshi VA, Li L, Yassin Y, Tamburino AM, Neel BG, Kucherlapati RS (January 2007). "Germline gain-of-function mutations in SOS1 cause Noonan syndrome". Nature Genetics. 39 (1): 70–4. doi:10.1038/ng1926. PMID 17143285. S2CID 10222262.
- Yoshikawa Y, Satoh T, Tamura T, Wei P, Bilasy SE, Edamatsu H, Aiba A, Katagiri K, Kinashi T, Nakao K, Kataoka T (August 2007). "The M-Ras-RA-GEF-2-Rap1 pathway mediates tumor necrosis factor-alpha dependent regulation of integrin activation in splenocytes". Molecular Biology of the Cell. 18 (8): 2949–59. doi:10.1091/mbc.E07-03-0250. PMC 1949361. PMID 17538012.