Neocentromere
Neocentromeres are new centromeres that form at a place on the chromosome that is usually not centromeric. They typically arise due to disruption of the normal centromere.[1] These neocentromeres should not be confused with “knobs”, which were also described as “neocentromeres” in maize in the 1950s.[2] Unlike most normal centromeres, neocentromeres do not contain satellite sequences that are highly repetitive but instead consist of unique sequences. Despite this, most neocentromeres are still able to carry out the functions of normal centromeres[1] in regulating chromosome segregation and inheritance. This raises many questions on what is necessary versus what is sufficient for constituting a centromere.
As neocentromeres are still a relatively new phenomenon in cell biology and genetics, it may be useful to keep in mind that neocentromeres may be somewhat related to point centromeres, holocentromeres, and regional centromeres. Whereas point centromeres are defined by sequence, regional and holocentromeres are epigenetically defined by where a specific type of nucleosome (the one containing the centromeric histone H3) is located.[3]
It may also be analytically helpful to take into account that the centromere is generally defined in relation to the kinetochore, specifically as the “part of the chromosome that links two sister chromatids together via the kinetochore”. However, the emergence of research in neocentromeres troubles this conventional definition and questions the function of a centromere beyond being a “landing pad” for kinetochore formation.[4] This expands the scope of the centromere's function to include regulating the function of the kinetochore and the mitotic spindle.
History
Neocentromeres were discovered relatively recently. They were first observed by Andy Choo in a human karyotype clinic case in 1997,[5] using fluorescent in situ hybridization (FISH) and cytogenetic analysis. The neocentromeres were observed on chromosome 10 of a patient, who was a child with developmental delays.
Cytogenetic and FISH analyses of his chromosomes found three marker chromosomes: one was a bisatellited chromosome, and two were derived from chromosome 10. Out of the two derived from chromosome 10, one formed a ring and the other a “deleted” version of chromosome 10, labelled as mar del(10).[5] The two derivations were confirmed to be from chromosome 10 using characterization of YACs and BACs;[6] this confirmation serves as a form of endogenous control for the neocentromere-instigating experiment. The mar del (10) and bisatellited chromosomes were present in every cell examined, but the ring chromosome was only present in 4–8% of the cells.[5] Some have attributed this statistic to the possible mitotic instability of ring chromosomes.[7]
Centromeres are conventionally defined by dark-staining heterochromatin which consists of the primary constriction site[7]—this convention exists because heterochromatin is usually found flanking the centromere. With this in mind, centromeric heterochromatin was found on the bisatellited and ring markers, but not on the mar del(10) marker. However, mar del(10) was still able to stably segregate in vivo and in vitro which implied the presence of a functional centromere and kinetochore. There has been an assumption that centromeric heterochromatin is just as important as the kinetochore in segregating and stabilizing chromosomes, because heterochromatin is associated with protein recruitment and has the defining ability to silence gene expression.[8] However, this notion is challenged by the observation in mar del (10).
A decade after the initial observation of neocentromeres, 60 more cases of human neocentromeres from across the genome, not just chromosome 10, were documented by 2002.[7] These cases were also typically observed in children with developmental delay or congenital abnormalities.[7] By 2012, more than 90 cases of human neocentromeres across 20 different chromosomes have been described.[9]
Formation
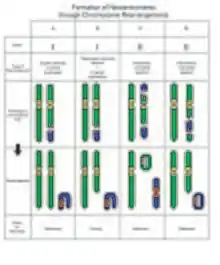
Studies suggest that neocentromeres are ultimately formed from epigenetic processes, rather than from changes in DNA sequence.[7] There is a general consensus that neocentromeres result from an attempt to fix chromosomes that lack a conventional centromere, through chromosome rearrangements.
The most common type of rearrangement leading to a neocentromere is an inverted duplication, categorized as Class I. The resulting marker chromosome consists of two copies of the chromosome segment. Each of the two copies is a mirror image to the chromosome segment. The neocentromere forms at an interstitial site, between the break point and one of the telomeres.[7] Despite having identical sequences in two halves of the chromosome, the neocentromere is only formed once. In some cases, segregation of these chromosomes result in partial trisomy, and at other times, a partial tetrasomy. In partial tetrasomy, the karyotype appears normal except for the marker chromosome.
Mechanisms of formation of neocentromeres are still unclear, but a few have been proposed. It is strongly speculated that neocentromeres form during mitosis or meiosis.[10]
For Class I, the proposed mechanism is that chromatid breakage occurs during mitosis, resulting in a chromosome fragment. That acentric chromosome fragment may segregate with the intact chromatid and result in partial tetrasomy; or, it may segregate with the complementary broken chromatid and result in partial trisomy because the broken chromatid may be saved by telomere restitution.[10] In both cases, the inverted duplicated marker forms only after cell division and replication by rejoining the broken, replicated ends of the fragment.[10] It is also suggested that U-type exchange during meiosis I may lead to partial tetrasomy.
On the other hand, Class II marker chromosomes result from the second most common type of rearrangement: interstitial deletions. A chromosome is rearranged to give a ring chromosome, and a linear chromosome. Hence, in hindsight, the first observation of neocentromeres made by Andy Choo in 1997 was most likely an example of a Class II pericentric interstitial deletion, followed by a complex rearrangement. The neocentromere can either appear on the linear chromosome or on the ring chromosome, depending on which ever one lacks a centromere.[10]
For Class II, it is unclear when the rearrangement occurs. General postulations include the chromosome breaking twice, with the ends rejoining. The alternative argument is that during meiosis I, looping and homologous recombination within a sister chromatid can cause this rearrangement.[10]
A rough estimation approximates that neocentromere formation on inverted duplicated chromosomes happens every 70,000–200,000 live births.[10] However, this statistic does not include Class II rearrangements.
Neocentromeres in humans and human diseases
Human centromeres usually consist of 2000–4000 kilobases of a 171 base pair repeat unit. This alpha satellite is not present at all in human neocentromeres.[10] By 2008, more than 90 reported cases of human neocentromeres were detected on marker chromosomes that experienced a loss of centromere, and a subsequent rearrangement.[10] Common traits among these human neocentromeres include being analphoid, staining C-band negative, containing a primary constriction site, and binding to essential centromere proteins that signal the presence of a kinetochore.[10]
Neocentromeres have been detected in human cancers,[10] even though the observed frequency of neocentromeres in the population is relatively low. There is a strong correlation between neocentromeres and a specific class (ALP-WDLPS) of lipomatous tumors.[11] It has been observed that lipomatous tumors with alphoid sequences in the centromere are more aggressive and metastatic. Hence, one suggestion is that neocentromeres may quickly evolve into a centromere with alpha satellite but the correlation between repetitive sequence and aggressiveness of tumors remains unclear.[10]
Neocentromeres have also been reported in lung carcinoma and acute myeloid leukemia.[10] It may be worth noting that neocentromeres in cancer may occur more frequently than currently observed, because many cancer screenings involving karyotyping do not use assays that detect neocentromere formations.
Although not discussed explicitly in relation to neocentromeres, it is worth noting that the breakage-fusion-bridge cycle involves ring chromosomes too.[12] Particularly, these ring chromosomes involved in breakage-fusion-bridge cycles have a relatively high percentage of prevalence in lipomatous tumors and a lower but still notable prevalence in lung and acute myelogenous leukemia, according to the Mitelman Database. Perhaps further studying of ring chromosomes and diseases, from the perspective of neocentromere formation, may shed light on mechanisms of tumor formation due to chromosome abnormalities. For example, one may question whether the ring chromosomes present in tumors contain a neocentromere following a Class II deletion, because the neocentromere may or may not be present on the ring.
As well as autosomes, human neocentromeres have also been observed in sex chromosomes, and correlate with some sex-linked diseases. In 1999, analysis of a neocentromere in the Y chromosome with a constriction site in the amniocytes of a 38-year-old woman, and in her husband and brother-in-law served as experimental evidence that neocentromeres can be stably transmitted through meiosis into the next generation.[13] Not only is the neocentromere viable on a chromosome, it is also sufficient for allowing proper male sex determination.[14] These meiotically transmitted neocentric markers show mosaicism, which many think are due to mitotic instability.[7] However, in contrary, others also think that mosaicism could be developed post-zygotically and hence may not be a result of mitotic instability.[5] Post-zygotic formation happens when the function of the neocentromere is not established at the time of meiotic rearrangement.
Another case of mosaicism has been observed in a 15-year-old girl with a neocentric X chromosome.[9] Her clinical features, along with cytogenetic and FISH analyses confirm that this is the first case of a mosaic Turner syndrome involving a neocentromere. During the time of this experiment, only two other cases of neocentric X chromosomes had been observed thus far, making this experiment the third. One of the two previous cases was also confirmed to be Turner syndrome but was not mosaic. An interaction of interest here would be between neocentromeres and the XIST gene, which is responsible for X-inactivation. It has been suggested that the abnormality caused by neocentromeres may account for the selective inactivation of the abnormal X chromosome in this patient. Taking into account that only fewer than 5% of Turner syndrome cases are mosaic,[9] one may consider, likewise with neocentromere assays in cancer,[10] that neocentromeres may occur at a higher frequency in mosaic Turner syndrome than observed.
Epigenetic regulation
As aforementioned, the formation of the centromere is well known to be regulated epigenetically. However, those epigenetic mechanisms are still debated contentiously; fortunately, neocentromeres provide a model system for studying different proposed mechanisms.[7][15]
Centromeric proteins
Centromeres are well-associated with specific proteins that are involved in the formation of the kinetochore and the mitotic spindle. Because neocentromeres do not contain repetitive sequences, they are good candidates for studying the epigenetic regulation of distributing centromeric proteins, using chromatin immunoprecipitation (ChIP) methods.[1]
All centromeres are associated with centromere protein A (CENPA).[8] CENPA also been widely studied as an important player in centromere regulation as it binds and specifies centromere location for both normal centromeres and neocentromeres, regardless of DNA sequence. There is a general consensus that CENPA assembles into octameric nucleosomes, with two copies of CENPA replacing two copies of histone H3 at the target locus[16]—this is the simplest model and much is still unknown about the composition of CENPA nucleosomes.
Neocentromeres have been useful tools in investigating the effects of CENPA in the absence of satellite sequences. Those studies conclude that the ability of CENPA to wrap DNA seem to be independent of DNA sequence.[16] This leads to several questions: how does CENPA, then, decide where to wrap? What is the purpose of satellite sequences (that are present at most eukaryotic centromeres) if they are not needed for CENPA wrapping? To complicate the matter even further, introducing alpha satellite DNA into cells can cause de novo formation of centromeres.[7] So far, this suggests that the repetitive sequences may play a fundamental but not necessary role in centromere formation. Moreover, non-repetitive centromeres have also been recently observed in horses, orangutans and chickens.[1][17]
Overexpression of CENPA and CENPH (centromere protein H) is also associated with colorectal cancer. It may be worth noticing that the overexpression of these centromeric proteins are also related to neocentromerization. Hence, this may serve as the start of the explanation to how neocentromeres may lead to cancer. Overexpression may be due to a loss of function in regulation of CENPA by proteolysis at inappropriate times of the cell cycle.[18] However, this link needs to be further investigated.[10]
Histone modifications
That neocentromeres and conventional centromeres do not share consistent chromatin environment should also be taken into account in questioning the epigenetic regulation of centromere formation.[19] The N-terminal tails of histones can be modified in several ways, including phosphorylation, acetylation, methylation and ubiquitination. Although certain histone modifications at centromeres seem to serve a purpose—for example, contributing to a higher order organization of chromatins in mouse centromeres[18]—conventional and neocentromeres share very few modifications but arguably still maintain the same function of a centromere.
Histone chaperones
Replenishment of CENPA every cycle, which is important for reestablishing centromere identity, is carried out by HJURP (Holliday Junction Recognition Protein), or Scm3 in fungi and CAL1 in Drosophila.[19] Tethering HJURP to a non-centromeric locus can give rise to a neocentromere, even after the disassociation of HJURP. There seems to be a co-evolutionary relationship between the Drosophila chaperone CAL1 and CENPA which accounts for species incompatibility—this is discussed more below.[20]
Cell cycle coordination with CENPA deposition
Whereas deposition of CENPA happens during S phase for S. cerevisiae, two pathways of CENPA deposition in S. pombe determine when CENPA is deposited, namely S phase and G2. In Arabidopsis thaliana, experiments suggest that CENPA deposition via a replication-independent mechanism in G2. For humans, the time seems to be during early G1.[18]
This temporal regulation is important as it reveals the composition of centromeric chromatin during the time of kinetochore assembly in mitosis. Perhaps reimaging the formation of neocentromeres from the perspective of the cell-cycle might reveal more about what type of regulation is necessary for centromere formation.
Evolution
One may expect, since the centromere plays such a significant role in chromosome segregation and general inheritance, that the centromere would be highly conserved, in sequence or in epigenetic regulation. However, even though the CENPA histone variant is in fact conserved, there is a surprisingly large amount of diversity in the organization of centromeric chromatin, across different lineages.[21] Another striking thing in centromere evolution is that while centromere function is conserved among all eukaryotes the underlying DNA is rapidly evolving, this phenomenon has been coined as the "centromere paradox".[22]
Yeast, Drosophila and mammals all have heterochromatin flanking their centromeres.[8] Although the vertebrate chaperone HJURP and the yeast chaperone Scm3 have diverged, their N-terminal domains show striking conservation.[23] On the other hand, frogs and chickens have domains in their chaperones that are not at all shared with that of yeast. Hence, further research into the mechanistic properties of these chaperones may potentially reveal how they help determine where and what type of centromere and neocentromere form.
Another element to consider from the evolutionary standpoint is that because neocentromeres are viable and can be transmitted meiotically from one generation to the next, they may play a role in species evolution. Recently, it was shown in Drosophila that co-evolution of CENPA and its chaperone CAL1 may explain species incompatibility.[20] This incompatibility exists between centromeric histones. This observation encourages neocentromeres to be studied along with their chaperones, to see if neocentromeres may also perhaps have “neo-chaperones” accompanying them.
Over time neocentromeres may also spread in a population and "mature" into full centromeres by accumulating repetitive elements, this can then lead to something known as evolutionary new centromeres.[24][25][26] Centromere repositioning and evolutionary new centromeres are also thought to be involved in speciation as incompatibility between centromeres may give rise to reproductive barriers.[27][28]
Related phenomena
Other phenomena or studies, that may or may not be explicitly related to neocentromeres, may pertain to connections that have not been made in scientific literature yet.
Holocentromeres, which are point centromeres distributed throughout the chromosome, have been most extensively studied in worms, C. elegans.[8] Holocentromeres serve as model comparisons to neocentromeres because holocentromeres have seemingly arbitrary “seeds” of CENPA distributed throughout the chromosome, which work together to make a functional kinetochore.[1] It is important to note that these CENPA seeds are excluded from genes or loci that are transcribed in the germline or early embryo. This leads to the thought that the seemingly random scattering of these seeds is not inherited, and that each generation or meiosis has its own distinct scatter.
Findings in model organisms, namely inducing neocentromerization in chicken and fungal systems, have brought up a few more correlations for thought.[19] Specifically, in chicken DT40 cells, it was found that neither histone modifications nor early replication timing is associated with neocentromere formation.[29] Moreover, it was also found that neocentromeres form on both transcriptionally active and inactive loci,[29] challenging the widely accepted notion that centromeres do not appear on coding regions of the chromosome. This leads to questions of how neocentromeres may disrupt transcription and expression of those genes.
Finally, a recent discovery is having double-stranded RNA specify centromere location.[8] It seems that the arrays of repetitive sequence flanking the centromere are transcribed into RNA that may in turn become RNAi machinery that aids the formation of heterochromatin. This may possibly relate to regulating the levels of centromeric proteins, similar to how CENPA levels are regulated by the cell cycle and proteolysis.
References
- Fukagawa, Tatsuo; Earnshaw, William C. (2014-10-06). "Neocentromeres". Current Biology. 24 (19): R946–R947. doi:10.1016/j.cub.2014.08.032. PMID 25291631.
- Rhoades, M. M.; Vilkomerson, H. (1942-10-01). "On the Anaphase Movement of Chromosomes". Proceedings of the National Academy of Sciences of the United States of America. 28 (10): 433–436. Bibcode:1942PNAS...28..433R. doi:10.1073/pnas.28.10.433. ISSN 0027-8424. PMC 1078510. PMID 16588574.
- Steiner, Florian A.; Henikoff, Steven (2014-01-01). "Holocentromeres are dispersed point centromeres localized at transcription factor hotspots". eLife. 3: e02025. doi:10.7554/elife.02025. ISSN 2050-084X. PMC 3975580. PMID 24714495.
- Verdaasdonk, Jolien S.; Bloom, Kerry (2011-05-01). "Centromeres: unique chromatin structures that drive chromosome segregation". Nature Reviews Molecular Cell Biology. 12 (5): 320–332. doi:10.1038/nrm3107. ISSN 1471-0080. PMC 3288958. PMID 21508988.
- Voullaire, L.; Saffery, R.; Earle, E.; Irvine, D. V.; Slater, H.; Dale, S.; du Sart, D.; Fleming, T.; Choo, K. H. (2001-07-22). "Mosaic inv dup(8p) marker chromosome with stable neocentromere suggests neocentromerization is a post-zygotic event". American Journal of Medical Genetics. 102 (1): 86–94. doi:10.1002/1096-8628(20010722)102:1<86::aid-ajmg1390>3.0.co;2-t. ISSN 0148-7299. PMID 11471179.
- Cancilla, M. R.; Tainton, K. M.; Barry, A. E.; Larionov, V.; Kouprina, N.; Resnick, M. A.; Sart, D. D.; Choo, K. H. (1998-02-01). "Direct cloning of human 10q25 neocentromere DNA using transformation-associated recombination (TAR) in yeast". Genomics. 47 (3): 399–404. doi:10.1006/geno.1997.5129. ISSN 0888-7543. PMID 9480754.
- Amor, David J.; Choo, K. H. Andy (2002-10-01). "Neocentromeres: Role in Human Disease, Evolution, and Centromere Study". American Journal of Human Genetics. 71 (4): 695–714. doi:10.1086/342730. ISSN 0002-9297. PMC 378529. PMID 12196915.
- Sullivan, B. A. (2013-01-01). "Centromeres". In Lane, M. Daniel (ed.). Centromeres A2 - Lennarz, William J. Waltham: Academic Press. pp. 446–450. doi:10.1016/B978-0-12-378630-2.00471-0. ISBN 9780123786319.
- Hemmat, Morteza; Wang, Boris T.; Warburton, Peter E.; Yang, Xiaojing; Boyar, Fatih Z.; El Naggar, Mohammed; Anguiano, Arturo (2012-01-01). "Neocentric X-chromosome in a girl with Turner-like syndrome". Molecular Cytogenetics. 5 (1): 29. doi:10.1186/1755-8166-5-29. ISSN 1755-8166. PMC 3477003. PMID 22682421.
- Marshall, Owen J.; Chueh, Anderly C.; Wong, Lee H.; Choo, K.H. Andy (2008-02-08). "Neocentromeres: New Insights into Centromere Structure, Disease Development, and Karyotype Evolution". American Journal of Human Genetics. 82 (2): 261–282. doi:10.1016/j.ajhg.2007.11.009. ISSN 0002-9297. PMC 2427194. PMID 18252209.
- Pedeutour, F; Forus, A; Coindre, J M; Berner, J M; Nicolo, G; Michiels, J F; Terrier, P; Ranchere-Vince, D; Collin, F (1999-02-01). "Structure of the supernumerary ring and giant rod chromosomes in adipose tissue tumors". Genes, Chromosomes and Cancer. 24 (1): 30–41. doi:10.1002/(SICI)1098-2264(199901)24:1<30::AID-GCC5>3.0.CO;2-P. ISSN 1045-2257. PMID 9892106.
- Gisselsson, D.; Höglund, Mattias; Mertens, Fredrik; Johansson, Bertil; Cin, Paola Dal; Berghe, Herman Van den; Earnshaw, William C.; Mitelman, Felix; Mandahl, Nils (1999-05-01). "The structure and dynamics of ring chromosomes in human neoplastic and non-neoplastic cells". Human Genetics. 104 (4): 315–325. doi:10.1007/s004390050960. ISSN 0340-6717. PMID 10369161.
- Tyler-Smith, C.; Gimelli, G.; Giglio, S.; Floridia, G.; Pandya, A.; Terzoli, G.; Warburton, P. E.; Earnshaw, W. C.; Zuffardi, O. (1999-05-01). "Transmission of a fully functional human neocentromere through three generations". American Journal of Human Genetics. 64 (5): 1440–1444. doi:10.1086/302380. ISSN 0002-9297. PMC 1377882. PMID 10205277.
- Velinov, M.; Gu, H.; Genovese, M.; Duncan, C.; Warburton, P.; Brooks, S. Sklower; Jenkins, E. C. (2004-06-01). "Characterization of an analphoid, neocentromere-positive inv dup 8p marker chromosome using multiplex whole chromosome and sub-telomere FISH analyses". Annales de Génétique. 47 (2): 199–205. doi:10.1016/j.anngen.2004.02.005. ISSN 0003-3995. PMID 15183754.
- Warburton, P. E.; Dolled, M.; Mahmood, R.; Alonso, A.; Li, S.; Naritomi, K.; Tohma, T.; Nagai, T.; Hasegawa, T. (2000-06-01). "Molecular cytogenetic analysis of eight inversion duplications of human chromosome 13q that each contain a neocentromere". American Journal of Human Genetics. 66 (6): 1794–1806. doi:10.1086/302924. ISSN 0002-9297. PMC 1378043. PMID 10777715.
- Hasson, Dan; Panchenko, Tanya; Salimian, Kevan J.; Salman, Mishah U.; Sekulic, Nikolina; Alonso, Alicia; Warburton, Peter E.; Black, Ben E. (2013-06-01). "The octamer is the major form of CENP-A nucleosomes at human centromeres". Nature Structural & Molecular Biology. 20 (6): 687–695. doi:10.1038/nsmb.2562. ISSN 1545-9993. PMC 3760417. PMID 23644596.
- Fukagawa, Tatsuo; Earnshaw, William C. (2014-09-08). "The Centromere: Chromatin Foundation for the Kinetochore Machinery". Developmental Cell. 30 (5): 496–508. doi:10.1016/j.devcel.2014.08.016. ISSN 1534-5807. PMC 4160344. PMID 25203206.
- Bernad, Rafael; Sánchez, Patricia; Losada, Ana (2009-11-15). "Epigenetic specification of centromeres by CENP-A". Experimental Cell Research. 315 (19): 3233–3241. doi:10.1016/j.yexcr.2009.07.023. PMID 19660450.
- Scott, Kristin C.; Sullivan, Beth A. (2014-02-01). "Neocentromeres: a place for everything and everything in its place". Trends in Genetics. 30 (2): 66–74. doi:10.1016/j.tig.2013.11.003. ISSN 0168-9525. PMC 3913482. PMID 24342629.
- Zasadzińska, Ewelina; Foltz, Daniel R. (2016-04-18). "Centromeres of a Different CAL-ibre". Developmental Cell. 37 (2): 105–106. doi:10.1016/j.devcel.2016.04.006. PMID 27093076.
- Steiner, Florian A.; Henikoff, Steven (2015-04-01). "Diversity in the organization of centromeric chromatin". Current Opinion in Genetics & Development. 31: 28–35. doi:10.1016/j.gde.2015.03.010. ISSN 1879-0380. PMID 25956076.
- Henikoff, S. (2001-08-10). "The Centromere Paradox: Stable Inheritance with Rapidly Evolving DNA". Science. 293 (5532): 1098–1102. doi:10.1126/science.1062939.
- Müller, Sebastian; Almouzni, Geneviève (2014-03-01). "A network of players in H3 histone variant deposition and maintenance at centromeres". Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms. 1839 (3): 241–250. doi:10.1016/j.bbagrm.2013.11.008. ISSN 0006-3002. PMID 24316467.
- Montefalcone, G. (1999-12-01). "Centromere Repositioning". Genome Research. 9 (12): 1184–1188. doi:10.1101/gr.9.12.1184. PMC 311001. PMID 10613840.
- Rocchi, M; Archidiacono, N; Schempp, W; Capozzi, O; Stanyon, R (January 2012). "Centromere repositioning in mammals". Heredity. 108 (1): 59–67. doi:10.1038/hdy.2011.101. ISSN 0018-067X. PMC 3238114. PMID 22045381.
- Tolomeo, Doron; Capozzi, Oronzo; Stanyon, Roscoe R.; Archidiacono, Nicoletta; D’Addabbo, Pietro; Catacchio, Claudia R.; Purgato, Stefania; Perini, Giovanni; Schempp, Werner; Huddleston, John; Malig, Maika (2017-02-03). "Epigenetic origin of evolutionary novel centromeres". Scientific Reports. 7 (1): 41980. doi:10.1038/srep41980. ISSN 2045-2322. PMC 5290474. PMID 28155877.
- Brown, Judith D.; O'Neill, Rachel J. (September 2010). "Chromosomes, Conflict, and Epigenetics: Chromosomal Speciation Revisited". Annual Review of Genomics and Human Genetics. 11 (1): 291–316. doi:10.1146/annurev-genom-082509-141554. ISSN 1527-8204.
- Hartley, Gabrielle; O’Neill, Rachel (2019-03-16). "Centromere Repeats: Hidden Gems of the Genome". Genes. 10 (3): 223. doi:10.3390/genes10030223. ISSN 2073-4425. PMC 6471113. PMID 30884847.
- Shang, Wei-Hao; Hori, Tetsuya; Martins, Nuno M. C.; Toyoda, Atsushi; Misu, Sadahiko; Monma, Norikazu; Hiratani, Ichiro; Maeshima, Kazuhiro; Ikeo, Kazuho (2013-03-25). "Chromosome engineering allows the efficient isolation of vertebrate neocentromeres". Developmental Cell. 24 (6): 635–648. doi:10.1016/j.devcel.2013.02.009. ISSN 1878-1551. PMC 3925796. PMID 23499358.