RHEB
RHEB also known as Ras homolog enriched in brain (RHEB) is a GTP-binding protein that is ubiquitously expressed in humans and other mammals. The protein is largely involved in the mTOR pathway and the regulation of the cell cycle.[5]
RHEB is a recently discovered member of the Ras superfamily. Being a relative of Ras, the overexpression of RHEB can be seen in multiple human carcinomas.[6] For this reason, ways to inhibit RHEB to control the mTOR pathway are studied as possible treatments for uncontrollable tumor cell growth in several diseases, especially in tuberous sclerosis.[7]
Structure
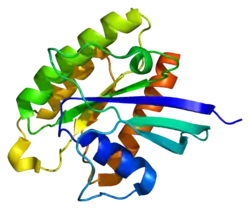
Rheb is a 21-kDa protein monomer composed of 184 amino acids.[5] The first 169 amino acids by the N-terminus make up the GTPase domain, and the remaining amino acids are part of a hypervariable region ending at the C-terminus in a CAAX motif (C – cysteine, A – aliphatic amino acid, X – C-terminus amino acid).[8]
The protein is a lipid-anchored, cell-membrane protein with five repeats of the RAS-related GTP-binding region.[5] Also present are “switch” regions, I and II, which undergo conformational changes when shuttling between GTP-bound(activated) and GDP-bound(inactive) forms.[8]
RHEB is expressed by the RHEB gene in humans.[9] Three pseudogenes have been mapped, two on chromosome 10 and one on chromosome 22.[5]
Function
Activation of mTORC1
RHEB is vital in regulation of growth and cell cycle progression due to its role in the insulin/TOR/S6K signaling pathway.[10] Mechanistic Target of Rapamycin Complex 1 (mTORC1) is a serine/threonine kinase whose activation leads to phosphorylation cascades within the cell that lead to cell growth and proliferation.[11] RHEB localizes at the lysosome to activate mTORC1 and Rag7 proteins localize mTORC1 to the lysosome and the Ragulator-Rag complex, allowing RHEB to activate the protein.[12] RHEB acts as an activator for mTORC1 in its GTP-bound form, therefore GTP-bound RHEB activates cell growth and proliferation within the cell.
mTORC1 independent functions
RHEB can serve as a regulator, for other proteins independent from mTORC1. For example, RHEB is an activator for nucleotide synthesis by binding carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase (CAD), an enzyme required for de novo pyrimidine nucleotide synthesis.[13] An increased nucleotide pool within the cell can lead to increased cell proliferation. mTORC1 is also a regulator for CAD, so both RHEB and mTORC1 are involved with the control of nucleotide level within the cell.[13] 5' adenosine-monophospate-activated protein kinase (AMPK) has also been found to be an effector for RHEB.[14] AMPK is a protein kinase that begins a phosphorylation cascade leading to autophagy. In rat studies, RHEB activates AMPK.[14] RHEB has also been found to interact with effectors upstream in the mTOR pathway. Phospholipase D1 (PLD1) is upstream in the mTOR pathway and serves as a positive effector for mTORC1.[15]
Other functions
RHEB may be involved in neural plasticity. This function is novel and not typically associated with the Ras proteins. Deficiency of RHEB in the forebrain of mice embryos is associated with decreased myelinization due to a decrease of mature oligodendrocytes.[8]
In studies of RHEB knockout mice, it was shown through hematoxylin-eosin staining that heart development is highly impaired. The cardiac myocytes do not sufficiently grow in size, indicating that RHEB mTOR function is required. This suggested that RHEB and the activation of the mTOR pathway is a necessity for proper cardiac development in mice embryos.[16]
Differences from Ras superfamily
RHEB functions differently compared to other proteins in the Ras superfamily.[8] Similar to those in the Ras superfamily, the protein has GTPase activity and shuttles between a GDP-bound form and a GTP-bound form, and farnesylation of the protein is required for this activity. However, unlike those in the Ras superfamily, conformational change when shuttling between forms only affects switch I, while switch II remains relatively stable, due to difference in secondary structure. Ras switch II forms a long α-helical structure between shuttling, while RHEB switch II adopts a more atypical conformation allowing for novel functions.[17] Such a conformation causes a decreased intrinsic rate of GTP hydrolysis as compared to RAS due to the catalytic Asp65 in the switch II region of RHEB being blocked from the active site.[11]
Regulation
GTP hydrolysis activity of RHEB is intrinsically slow and the GTP-bound form is more common, thus RHEB is more likely active than not active within the cell.[11] Its activity is strongly regulated within the cell by tumor-suppressant proteins that form the TSC complex. Specifically, the TSC2 subunit, tuberin of the complex interacts with and inhibits RHEB to regulate the protein. Tuberin stimulates RHEB to hydrolyze GTP, thus inactivating it.[18]
Tuberous sclerosis
Tuberous sclerosis is an autosomal dominant disease in which the genes required to express the tumor-suppressant proteins that form the TSC complex is mutated or missing, so the TSC complex is unable to function properly.[19] This could lead to the disregulation of many signalling proteins and effectors within the cell, including RHEB. Unregulated activity of RHEB can lead to uncontrollable cell growth and cell division which could ultimately lead to formation of tumors.[8]
Interactions
RHEB has been shown to interact with:
- Ataxia telangiectasia mutated (ATM)[20]
- Ataxia telangiectasia and Rad3 related (ATR)[20]
- 5' AMP-activated protein kinase (AMPK)[14]
- RAF proto-oncogene serine/threonine-protein kinase (C-Raf)[20][21][22]
- mammalian Target of Rapamycin Complex 1 (mTORC1),[20][23][24][25]
- Phospholipase D1 (PLD1)[15]
- Regulatory-associated protein of mTOR (RPTOR)[20]
- Tuberous sclerosis complex (TSC)[18][20][26][27][28][29] and
- Carbamoyl-phosphate synthetase 2, aspartate transcarbamoylase, dihydroorotase (CAD)[13]
References
- GRCh38: Ensembl release 89: ENSG00000106615 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000028945 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "RHEB Ras homolog enriched in brain [Homo sapiens (human)]". Gene - NCBI. National Center for Biotechnology Information, United States National Institutes of Health.
- Lu ZH, Shvartsman MB, Lee AY, Shao JM, Murray MM, Kladney RD, Fan D, Krajewski S, Chiang GG, Mills GB, Arbeit JM (Apr 2010). "Mammalian target of rapamycin activator RHEB is frequently overexpressed in human carcinomas and is critical and sufficient for skin epithelial carcinogenesis". Cancer Research. 70 (8): 3287–98. doi:10.1158/0008-5472.CAN-09-3467. PMC 2855737. PMID 20388784.
- Sugiura H, Yasuda S, Katsurabayashi S, Kawano H, Endo K, Takasaki K, Iwasaki K, Ichikawa M, Kobayashi T, Hino O, Yamagata K (2015-01-01). "Rheb activation disrupts spine synapse formation through accumulation of syntenin in tuberous sclerosis complex". Nature Communications. 6: 6842. doi:10.1038/ncomms7842. PMID 25880340.
- Heard JJ, Fong V, Bathaie SZ, Tamanoi F (Sep 2014). "Recent progress in the study of the Rheb family GTPases". Cellular Signalling. 26 (9): 1950–7. doi:10.1016/j.cellsig.2014.05.011. PMC 4134338. PMID 24863881.
- Mizuki N, Kimura M, Ohno S, Miyata S, Sato M, Ando H, Ishihara M, Goto K, Watanabe S, Yamazaki M, Ono A, Taguchi S, Okumura K, Nogami M, Taguchi T, Ando A, Inoko H (May 1996). "Isolation of cDNA and genomic clones of a human Ras-related GTP-binding protein gene and its chromosomal localization to the long arm of chromosome 7, 7q36". Genomics. 34 (1): 114–8. doi:10.1006/geno.1996.0248. PMID 8661031.
- Patel PH, Thapar N, Guo L, Martinez M, Maris J, Gau CL, Lengyel JA, Tamanoi F (Sep 2003). "Drosophila Rheb GTPase is required for cell cycle progression and cell growth". Journal of Cell Science. 116 (Pt 17): 3601–10. doi:10.1242/jcs.00661. PMID 12893813.
- Mazhab-Jafari MT, Marshall CB, Ishiyama N, Ho J, Di Palma V, Stambolic V, Ikura M (Sep 2012). "An autoinhibited noncanonical mechanism of GTP hydrolysis by Rheb maintains mTORC1 homeostasis". Structure. 20 (9): 1528–39. doi:10.1016/j.str.2012.06.013. PMID 22819219.
- Groenewoud MJ, Zwartkruis FJ (Aug 2013). "Rheb and Rags come together at the lysosome to activate mTORC1". Biochemical Society Transactions. 41 (4): 951–5. doi:10.1042/BST20130037. PMID 23863162.
- Sato T, Akasu H, Shimono W, Matsu C, Fujiwara Y, Shibagaki Y, Heard JJ, Tamanoi F, Hattori S (Jan 2015). "Rheb protein binds CAD (carbamoyl-phosphate synthetase 2, aspartate transcarbamoylase, and dihydroorotase) protein in a GTP- and effector domain-dependent manner and influences its cellular localization and carbamoyl-phosphate synthetase (CPSase) activity". The Journal of Biological Chemistry. 290 (2): 1096–105. doi:10.1074/jbc.M114.592402. PMC 4294477. PMID 25422319.
- Lacher MD, Pincheira R, Zhu Z, Camoretti-Mercado B, Matli M, Warren RS, Castro AF (Dec 2010). "Rheb activates AMPK and reduces p27Kip1 levels in Tsc2-null cells via mTORC1-independent mechanisms: implications for cell proliferation and tumorigenesis". Oncogene. 29 (50): 6543–56. doi:10.1038/onc.2010.393. PMID 20818424. S2CID 205531885.
- Sun Y, Fang Y, Yoon MS, Zhang C, Roccio M, Zwartkruis FJ, Armstrong M, Brown HA, Chen J (Jun 2008). "Phospholipase D1 is an effector of Rheb in the mTOR pathway". Proceedings of the National Academy of Sciences of the United States of America. 105 (24): 8286–91. doi:10.1073/pnas.0712268105. PMC 2448829. PMID 18550814.
- Tamai T, Yamaguchi O, Hikoso S, Takeda T, Taneike M, Oka T, Oyabu J, Murakawa T, Nakayama H, Uno Y, Horie K, Nishida K, Sonenberg N, Shah AM, Takeda J, Komuro I, Otsu K (Apr 2013). "Rheb (Ras homologue enriched in brain)-dependent mammalian target of rapamycin complex 1 (mTORC1) activation becomes indispensable for cardiac hypertrophic growth after early postnatal period". The Journal of Biological Chemistry. 288 (14): 10176–87. doi:10.1074/jbc.M112.423640. PMC 3617260. PMID 23426372.
- Aspuria PJ, Tamanoi F (Oct 2004). "The Rheb family of GTP-binding proteins". Cellular Signalling. 16 (10): 1105–12. doi:10.1016/j.cellsig.2004.03.019. PMID 15240005.
- Castro AF, Rebhun JF, Clark GJ, Quilliam LA (Aug 2003). "Rheb binds tuberous sclerosis complex 2 (TSC2) and promotes S6 kinase activation in a rapamycin- and farnesylation-dependent manner". The Journal of Biological Chemistry. 278 (35): 32493–6. doi:10.1074/jbc.C300226200. PMID 12842888.
- Cheadle JP, Reeve MP, Sampson JR, Kwiatkowski DJ (Aug 2000). "Molecular genetic advances in tuberous sclerosis". Human Genetics. 107 (2): 97–114. doi:10.1007/s004390000348. PMID 11030407. S2CID 10960505.
- Long X, Lin Y, Ortiz-Vega S, Yonezawa K, Avruch J (Apr 2005). "Rheb binds and regulates the mTOR kinase". Current Biology. 15 (8): 702–13. doi:10.1016/j.cub.2005.02.053. PMID 15854902. S2CID 3078706.
- Karbowniczek M, Cash T, Cheung M, Robertson GP, Astrinidis A, Henske EP (Jul 2004). "Regulation of B-Raf kinase activity by tuberin and Rheb is mammalian target of rapamycin (mTOR)-independent". The Journal of Biological Chemistry. 279 (29): 29930–7. doi:10.1074/jbc.M402591200. PMID 15150271.
- Yee WM, Worley PF (Feb 1997). "Rheb interacts with Raf-1 kinase and may function to integrate growth factor- and protein kinase A-dependent signals". Molecular and Cellular Biology. 17 (2): 921–33. doi:10.1128/mcb.17.2.921. PMC 231818. PMID 9001246.
- Long X, Ortiz-Vega S, Lin Y, Avruch J (Jun 2005). "Rheb binding to mammalian target of rapamycin (mTOR) is regulated by amino acid sufficiency". The Journal of Biological Chemistry. 280 (25): 23433–6. doi:10.1074/jbc.C500169200. PMID 15878852.
- Smith EM, Finn SG, Tee AR, Browne GJ, Proud CG (May 2005). "The tuberous sclerosis protein TSC2 is not required for the regulation of the mammalian target of rapamycin by amino acids and certain cellular stresses". The Journal of Biological Chemistry. 280 (19): 18717–27. doi:10.1074/jbc.M414499200. PMID 15772076.
- Bernardi R, Guernah I, Jin D, Grisendi S, Alimonti A, Teruya-Feldstein J, Cordon-Cardo C, Simon MC, Rafii S, Pandolfi PP (Aug 2006). "PML inhibits HIF-1alpha translation and neoangiogenesis through repression of mTOR". Nature. 442 (7104): 779–85. doi:10.1038/nature05029. PMID 16915281. S2CID 4427427.
- Inoki K, Li Y, Xu T, Guan KL (Aug 2003). "Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling". Genes & Development. 17 (15): 1829–34. doi:10.1101/gad.1110003. PMC 196227. PMID 12869586.
- Garami A, Zwartkruis FJ, Nobukuni T, Joaquin M, Roccio M, Stocker H, Kozma SC, Hafen E, Bos JL, Thomas G (Jun 2003). "Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2" (PDF). Molecular Cell. 11 (6): 1457–66. doi:10.1016/s1097-2765(03)00220-x. PMID 12820960.
- Zhang Y, Gao X, Saucedo LJ, Ru B, Edgar BA, Pan D (Jun 2003). "Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins". Nature Cell Biology. 5 (6): 578–81. doi:10.1038/ncb999. PMID 12771962. S2CID 13451385.
- Cao Y, Kamioka Y, Yokoi N, Kobayashi T, Hino O, Onodera M, Mochizuki N, Nakae J (Dec 2006). "Interaction of FoxO1 and TSC2 induces insulin resistance through activation of the mammalian target of rapamycin/p70 S6K pathway". The Journal of Biological Chemistry. 281 (52): 40242–51. doi:10.1074/jbc.M608116200. PMID 17077083.
Further reading
- Yamagata K, Sanders LK, Kaufmann WE, Yee W, Barnes CA, Nathans D, Worley PF (Jun 1994). "rheb, a growth factor- and synaptic activity-regulated gene, encodes a novel Ras-related protein". The Journal of Biological Chemistry. 269 (23): 16333–9. PMID 8206940.
- Gromov PS, Madsen P, Tomerup N, Celis JE (Dec 1995). "A novel approach for expression cloning of small GTPases: identification, tissue distribution and chromosome mapping of the human homolog of rheb". FEBS Letters. 377 (2): 221–6. doi:10.1016/0014-5793(95)01349-0. PMID 8543055. S2CID 23656670.
- Bonaldo MF, Lennon G, Soares MB (Sep 1996). "Normalization and subtraction: two approaches to facilitate gene discovery". Genome Research. 6 (9): 791–806. doi:10.1101/gr.6.9.791. PMID 8889548.
- Clark GJ, Kinch MS, Rogers-Graham K, Sebti SM, Hamilton AD, Der CJ (Apr 1997). "The Ras-related protein Rheb is farnesylated and antagonizes Ras signaling and transformation". The Journal of Biological Chemistry. 272 (16): 10608–15. doi:10.1074/jbc.272.16.10608. PMID 9099708.
- Inohara N, Ding L, Chen S, Núñez G (Apr 1997). "harakiri, a novel regulator of cell death, encodes a protein that activates apoptosis and interacts selectively with survival-promoting proteins Bcl-2 and Bcl-X(L)". The EMBO Journal. 16 (7): 1686–94. doi:10.1093/emboj/16.7.1686. PMC 1169772. PMID 9130713.
- Sanger Centre, The; Washington University Genome Sequencing Cente, The (Nov 1998). "Toward a complete human genome sequence". Genome Research. 8 (11): 1097–108. doi:10.1101/gr.8.11.1097. PMID 9847074.
- Kita K, Wu YP, Sugaya S, Moriya T, Nomura J, Takahashi S, Yamamori H, Nakajima N, Suzuki N (Aug 2000). "Search for UV-responsive genes in human cells by differential mRNA display: involvement of human ras-related GTP-binding protein, Rheb, in UV susceptibility". Biochemical and Biophysical Research Communications. 274 (3): 859–64. doi:10.1006/bbrc.2000.3220. PMID 10924367.
- Hanzal-Bayer M, Renault L, Roversi P, Wittinghofer A, Hillig RC (May 2002). "The complex of Arl2-GTP and PDE delta: from structure to function". The EMBO Journal. 21 (9): 2095–106. doi:10.1093/emboj/21.9.2095. PMC 125981. PMID 11980706.
- Tabancay AP, Gau CL, Machado IM, Uhlmann EJ, Gutmann DH, Guo L, Tamanoi F (Oct 2003). "Identification of dominant negative mutants of Rheb GTPase and their use to implicate the involvement of human Rheb in the activation of p70S6K". The Journal of Biological Chemistry. 278 (41): 39921–30. doi:10.1074/jbc.M306553200. PMID 12869548.
- Tee AR, Manning BD, Roux PP, Cantley LC, Blenis J (Aug 2003). "Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb". Current Biology. 13 (15): 1259–68. doi:10.1016/S0960-9822(03)00506-2. PMID 12906785. S2CID 6519150.
External links
- RHEB+protein,+human at the US National Library of Medicine Medical Subject Headings (MeSH)
This article incorporates text from the United States National Library of Medicine, which is in the public domain.