Spanish flu research
Spanish flu research concerns studies regarding the causes and characteristics of the "Spanish flu," a variety of influenza that in 1918 was responsible for the worst influenza pandemic in modern history. Many theories about the origins and progress of the Spanish flu persisted in the literature, but it was not until 2005, when various samples of lung tissue were recovered from American World War I soldiers and from an Inupiat woman buried in permafrost in a mass grave in Brevig Mission, Alaska, that significant genetic research was made possible.
| Influenza A virus subtype H1N1 | |
|---|---|
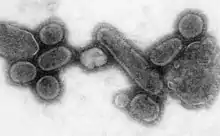 | |
| An electron micrograph of the virus that caused the 1918 flu. | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Kingdom: | Orthornavirae |
| Phylum: | Negarnaviricota |
| Class: | Insthoviricetes |
| Order: | Articulavirales |
| Family: | Orthomyxoviridae |
| Genus: | Alphainfluenzavirus |
| Species: | Influenza A virus |
| Serotype: | Influenza A virus subtype H1N1 |
| Sampled strains | |
| |
Origin of virus
There are two prevailing theories usually postulated. One theory by Alfred W. Crosby is that the virus strain originated at Fort Riley, Kansas, by two genetic mechanisms – genetic drift and antigenic shift – in viruses in poultry and swine which the fort bred for local consumption. Though initial data from a recent reconstruction of the virus suggested that it jumped directly from birds to humans, without traveling through swine,[lower-alpha 1] this has since been cast into doubt. One researcher published in 2004 argued that the disease was found in Haskell County, Kansas, as early as January 1918.[2] A similar and even more deadly virus had been seen earlier at British camps in France and at Aldershot.[3]
Earlier investigative work published in 2000 by a team led by British virologist, John Oxford[4] of St Bartholomew's Hospital and the Royal London Hospital, suggested that a principal British troop staging camp in Étaples, France, was at the center of the 1918 flu pandemic or at least a significant precursor virus to it. There had been a mysterious respiratory infection at the military base during the winter of 1915–1916.[5]
Discovery of viral genomes


In 1995, Jeffery Taubenberger of the US Armed Forces Institute of Pathology (AFIP), wondered if it might be possible to recover the virus of 1918 flu pandemic from the dried and fixed tissue of victims. He and his colleagues, tested 10 slides of tissue sample and 2 came out positive. Taubenberger, Ann H. Reid and Thomas G. Fanning were able to amplify short segments of the viral nucleic acid using polymerase chain reaction (PCR).[6] The results were published in the journal Science in March 1997.[7]
On Aug. 20, 1997, Johan Hultin recovered samples of the 1918 influenza from the frozen corpse of a Native Alaskan woman buried for nearly eight decades in permafrost near Brevig Mission, Alaska.[8] He brought the samples to a team in Rockville, Maryland led by Jeffery Taubenberger of the US Armed Forces Institute of Pathology (AFIP). Brevig Mission lost approximately 85% of its population to the 1918 flu in November 1918. One of the four recovered samples contained viable genetic material of the virus. This sample provided scientists a first-hand opportunity to study the virus, which was inactivated with guanidinium thiocyanate before transport. This sample and others found in AFIP archives allowed researchers to completely analyze the critical gene structures of the 1918 virus.
- "We have now identified three cases: The Brevig Mission case and two archival cases that represent the only known sources of genetic material of the 1918 influenza virus," said Taubenberger, chief of AFIP's molecular pathology division and principal investigator on the project.[9][lower-alpha 2]
The archived autopsy samples had been taken from WWI Army privates Roscoe Vaughan and James Downs.[11]
| Wikinews has related news: |
The 6 February 2004 edition of Science magazine reported that two research teams, one led by Sir John Skehel, director of the National Institute for Medical Research in London, another by professor Ian Wilson of The Scripps Research Institute in San Diego, had managed to synthesize the hemagglutinin protein responsible for the flu outbreak of 1918. They did this by piecing together DNA from a lung sample from an Inuit woman buried in the Alaskan tundra and a number of preserved samples from American soldiers of the First World War. The teams had analyzed the structure of the gene and discovered how subtle alterations to the shape of a protein molecule had allowed it to move from birds to humans with such devastating effects.
On 5 October 2005, Tumpey and other researchers at the Centers for Disease Control and Prevention (CDC) in Atlanta, Georgia, and the Mount Sinai School of Medicine in New York, announced that the (~13 kbp) genetic sequence of the 1918 flu strain, a subtype of avian strain H1N1, had been reconstructed using historic tissue samples and a small part of the RNA from a modern strain.[12][13][14]
Characteristics of virus
Influenza viruses have a relatively high mutation rate that is characteristic of RNA viruses. The H5N1 virus has mutated into a variety of types with differing pathogenic profiles; some pathogenic to one species but not others, some pathogenic to multiple species.[15] The ability of various influenza strains to show species-selectivity is largely due to variation in the hemagglutinin genes. Genetic mutations in the hemagglutinin gene that cause single amino acid substitutions can significantly alter the ability of viral hemagglutinin proteins to bind to receptors on the surface of host cells. Such mutations in avian H5N1 viruses can change virus strains from being inefficient at infecting human cells to being as efficient in causing human infections as more common human influenza virus types.[16]
In July 2004, researchers led by H. Deng of the Harbin Veterinary Research Institute, Harbin, China, and Robert Webster of the St. Jude Children's Research Hospital, Memphis, Tennessee, reported results of experiments in which mice had been exposed to 21 isolates of confirmed H5N1 strains obtained from ducks in China between 1999 and 2002. They found "a clear temporal pattern of progressively increasing pathogenicity."[17] Results reported by Webster in July 2005 reveal further progression toward pathogenicity in mice and longer virus shedding by ducks.
In December 2008, research by Yoshihiro Kawaoka of University of Wisconsin showed the presence of the three specific genes (termed PA, PB1, and PB2) and a nucleoprotein derived from the H1N1 1918 flu samples was enough to trigger similar symptoms in animal testing.[18]
Research of viral pathogenesis
Recent research of Taubenberger et al. has suggested that the 1918 virus, like H5N1, could have arisen directly from an avian influenza virus.[13] However, researchers at University of Virginia and Australian National University have suggested that there may be an alternative interpretation of the data used in the Taubenberger et al. paper.[19][20] Taubenberger et al. responded to these letters and defended their original interpretation.[21]
Other research by Tumpey and colleagues who reconstructed the H1N1 virus of 1918 came to the conclusion that it was most notably the polymerase genes and the HA and NA genes that caused the extreme virulence of this virus.[14] On 18 January 2007, Kobasa et al. reported that infected monkeys (Macaca fascicularis) exhibited classic symptoms of the 1918 pandemic and died from a cytokine storm.[22]
The sequences of the polymerase proteins (PA, PB1, and PB2) of the 1918 virus and subsequent human viruses differ by only 10 amino acids from the avian influenza viruses. Viruses with 7 of the 10 amino acids in the human influenza locations have already been identified in currently circulating H5N1. This has led some researchers to suggest that other mutations may surface and make the H5N1 virus capable of human-to-human transmission.
Another important factor is the change of the HA protein to a binding preference for alpha-2,6 sialic acid (the major form found in the human respiratory tract). In avian virus the HA protein preferentially binds to alpha-2,3 sialic acid, which is the major form in the avian enteric tract. It has been shown that only a single amino acid change can result in the change of this binding preference. Altogether, only a handful of mutations may need to take place in order for H5N1 avian flu to become a pandemic virus like the one of 1918. However it is important to note that likelihood of mutation does not indicate the likelihood for the evolution of such a strain, since some of the necessary mutations may be constrained by stabilizing selection.
Blood plasma as an effective treatment
In the event of another pandemic, US military researchers have proposed reusing a treatment from the deadly pandemic of 1918 in order to blunt the effects of the flu: Some military doctors injected severely afflicted patients with blood or blood plasma from people who had recovered from the flu. Data collected during that time indicates that the blood-injection treatment reduced mortality rates by as much as 50 percent.[23]
Navy researchers have launched a test to see if the 1918 treatment will work against deadly Asian bird flu. Results thus far have been inconclusive. Human H5N1 plasma may be an effective, timely, and widely available treatment for the next flu pandemic. A new international study using modern data collection methods, would be a difficult, slow process. Citing the months-long wait for a vaccine for the next pandemic, many flu experts are of the opinion that the 1918 method is something to consider.[24]
In the worldwide 1918 flu pandemic, "physicians tried everything they knew, everything they had ever heard of, from the ancient art of bleeding patients, to administering oxygen, to developing new vaccines and sera (chiefly against what we now call Hemophilus influenzae – a name derived from the fact that it was originally considered the etiological agent – and several types of pneumococci). Only one therapeutic measure showed any hint of success: Transfusing blood from recovered patients to new victims."[25]
See also
- Influenza research
- Mark Sykes – exhumation of a British flu victim in the United Kingdom
- Yoshihiro Kawaoka – engineered and recreated a virus to study how it works and how the flu naturally mutates
Footnotes
- Sometimes a virus contains both avian adapted genes and human adapted genes. Both the H2N2 and H3N2 pandemic strains contained avian flu virus RNA segments. "While the pandemic human influenza viruses of 1957 (H2N2) and 1968 (H3N2) clearly arose through reassortment between human and avian viruses, the influenza virus causing the 'Spanish Flu' in 1918 appears to be entirely derived from an avian source." (Belshe, 2005)[1]
- Johan Hultin first attempted to recover samples from Brevig in 1951, but was unsuccessful. In 1997, by then a seventy-two-year-old retired pathologist, Hultin decided that science had advanced enough to make another attempt worthwhile. Taubenberger had already recovered RNA of limited quality from samples of two servicemen who had died in the pandemic, and Hultin wrote offering his services to try to get better quality samples from Brevig permafrost. Taubenberger accepted, and Hultin went alone to Brevig in August 1997, and recovered the sample from the Alaskan woman, which Taubenberger and his team then analysed.[10]
References
- Harder, Timm C.; Werner, Ortrud (2006). "Chapter Two: Avian Influenza" (free on-line book). Influenza Report (Report).
a medical textbook that provides a comprehensive overview of epidemic and pandemic influenza.
- Barry, J.M. (January 2004). "The site of origin of the 1918 influenza pandemic and its public health implications". J Transl Med. 2 (1): 3. doi:10.1186/1479-5876-2-3. PMC 340389. PMID 14733617.
- "Origins of the 1918 pandemic: The case for France". National Public Radio (NPR). 5222069.
- "Dr. John Oxford". EU Research Profile. Archived from the original on 12 May 2009.
- Connor, Steve (8 January 2000). "Flu epidemic traced to Great War transit camp". The Independent. UK. Archived from the original on 8 August 2009. Retrieved 26 March 2020.
An investigation into the global epidemic of influenza in 1918, which killed an estimated 40 million people, has shown it almost certainly started in an army camp in France in the middle of the First World War.
- "Resurrecting 1918 Flu Virus Took Many Turns". 2005-10-10. ISSN 0190-8286. Retrieved 2020-07-25.
- Taubenberger, Jeffery K.; Reid, Ann H.; Krafft, Amy E.; Bijwaard, Karen E.; Fanning, Thomas G. (1997-03-21). "Initial Genetic Characterization of the 1918 "Spanish" Influenza Virus". Science. 275 (5307): 1793–1796. doi:10.1126/science.275.5307.1793. ISSN 0036-8075. PMID 9065404. S2CID 8976173.
- Brown, David (10 October 2005). "Resurrecting 1918 flu virus took many turns". Washington Post.
- "Lethal secrets of 1918 flu virus". BBC News.
- Kolata, Gina (1999). Flu: The story of the great influenza pandemic of 1918 and the search for the virus that caused it. Farrar, Straus, and Giroux. pp. 255–265. ISBN 978-0-374-15706-7.
- "Closing in on a killer: Scientists unlock clues to the Spanish influenza virus". National Museum of Health and Medicine. Retrieved 23 March 2020.
- "The 1918 flu virus is resurrected". Nature. 437 (7060): 794–795. October 2005. Bibcode:2005Natur.437..794.. doi:10.1038/437794a. PMC 7095040. PMID 16208326.
- Taubenberger JK, Reid AH, Lourens RM, Wang R, Jin G, Fanning TG (October 2005). "Characterization of the 1918 influenza virus polymerase genes". Nature. 437 (7060): 889–893. Bibcode:2005Natur.437..889T. doi:10.1038/nature04230. PMID 16208372. S2CID 4405787.
- Tumpey TM, Basler CF, Aguilar PV, et al. (October 2005). "Characterization of the reconstructed 1918 Spanish influenza pandemic virus". Science. 310 (5745): 77–80. Bibcode:2005Sci...310...77T. CiteSeerX 10.1.1.418.9059. doi:10.1126/science.1119392. PMID 16210530. S2CID 14773861.
- Kou Z, Lei FM, Yu J, Fan ZJ, Yin ZH, Jia CX, Xiong KJ, Sun YH, Zhang XW, Wu XM, Gao XB, Li TX (2005). "New genotype of avian influenza H5N1 viruses isolated from tree sparrows in China". J. Virol. 79 (24): 15460–15466. doi:10.1128/JVI.79.24.15460-15466.2005. PMC 1316012. PMID 16306617.
- Gambaryan A, Tuzikov A, Pazynina G, Bovin N, Balish A, Klimov A (January 2006). "Evolution of the receptor binding phenotype of influenza A (H5) viruses". Virology. 344 (2): 432–438. doi:10.1016/j.virol.2005.08.035. PMID 16226289.
- Chen H, Deng G, Li Z, et al. (July 2004). "The evolution of H5N1 influenza viruses in ducks in southern China". Proc. Natl. Acad. Sci. U.S.A. 101 (28): 10452–10457. Bibcode:2004PNAS..10110452C. doi:10.1073/pnas.0403212101. PMC 478602. PMID 15235128.
- "Researchers unlock secrets of 1918 flu pandemic". Reuters. 29 December 2008.
- Gibbs MJ, Gibbs AJ (April 2006). "Molecular virology: Was the 1918 pandemic caused by a bird flu?". Nature. 440 (7088): E8, discussion E9–10. Bibcode:2006Natur.440E...8G. doi:10.1038/nature04823. PMID 16641948. S2CID 4422599.
- Antonovics J, Hood ME, Baker CH (April 2006). "Molecular virology: Was the 1918 flu avian in origin?". Nature. 440 (7088): E9, discussion E9–10. Bibcode:2006Natur.440E...9A. doi:10.1038/nature04824. PMID 16641950. S2CID 4382489.
- Taubenberger JK, Reid AH, Lourens RM, Wang R, Jin G, Fanning TG (April 2006). "Molecular virology: Was the 1918 pandemic caused by a bird flu? Was the 1918 flu avian in origin? (Reply)". Nature. 440 (7088): E9–E10. Bibcode:2006Natur.440E...9T. doi:10.1038/nature04825. S2CID 4308033.
- Kobasa D, Jones SM, Shinya K, Kash JC, Copps J, Ebihara H, et al. (January 2007). "Aberrant innate immune response in lethal infection of macaques with the 1918 influenza virus". Nature. 445 (7125): 319–323. Bibcode:2007Natur.445..319K. doi:10.1038/nature05495. PMID 17230189. S2CID 4431644.
- "Influenza (main)". history.navy.mil.
- "[no title cited]". National Public Radio (NPR). 5731688.
- "The Threat of Pandemic Influenza: Are we ready?" (free online book). www.ncbi.nlm.nih.gov. Workshop Summary. National Institutes of Health. 2005. p. 62.