Influenza A virus
Influenza A virus causes influenza in birds and some mammals, and is the only species of the genus Alphainfluenzavirus of the virus family Orthomyxoviridae.[1] Strains of all subtypes of influenza A virus have been isolated from wild birds, although disease is uncommon. Some isolates of influenza A virus cause severe disease both in domestic poultry and, rarely, in humans.[2] Occasionally, viruses are transmitted from wild aquatic birds to domestic poultry, and this may cause an outbreak or give rise to human influenza pandemics.[3][4]
| Influenza A virus | |
|---|---|
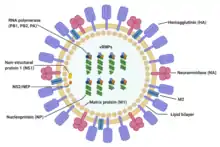 | |
| Structure of influenza A virus | |
 | |
| TEM micrograph of influenza A viruses | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Kingdom: | Orthornavirae |
| Phylum: | Negarnaviricota |
| Class: | Insthoviricetes |
| Order: | Articulavirales |
| Family: | Orthomyxoviridae |
| Genus: | Alphainfluenzavirus |
| Species: | Influenza A virus |
| Subtypes | |
Influenza A viruses are negative-sense, single-stranded, segmented RNA viruses. The several subtypes are labeled according to an H number (for the type of hemagglutinin) and an N number (for the type of neuraminidase). There are 18 different known H antigens (H1 to H18) and 11 different known N antigens (N1 to N11).[5][6] H17N10 was isolated from fruit bats in 2012.[7][8] H18N11 was discovered in a Peruvian bat in 2013.[6]
Each virus subtype has mutated into a variety of strains with differing pathogenic profiles; some are pathogenic to one species but not others, some are pathogenic to multiple species.
A filtered and purified influenza A vaccine for humans has been developed and many countries have stockpiled it to allow a quick administration to the population in the event of an avian influenza pandemic. Avian influenza is sometimes called avian flu, and colloquially, bird flu. In 2011, researchers reported the discovery of an antibody effective against all types of the influenza A virus.[9]
Variants and subtypes
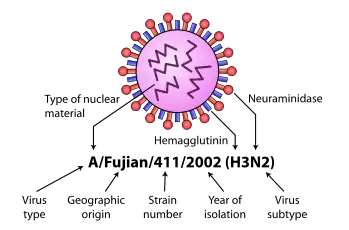
Influenza type A viruses are RNA viruses categorized into subtypes based on the type of two proteins on the surface of the viral envelope:
- H = hemagglutinin, a protein that causes red blood cells to agglutinate.
- N = neuraminidase, an enzyme that cleaves the glycosidic bonds of the monosaccharide sialic acid (previously called neuraminic acid).
The hemagglutinin is central to the virus's recognizing and binding to target cells, and also to its then infecting the cell with its RNA. The neuraminidase, on the other hand, is critical for the subsequent release of the daughter virus particles created within the infected cell so they can spread to other cells.
Different influenza viruses encode for different hemagglutinin and neuraminidase proteins. For example, the H5N1 virus designates an influenza A subtype that has a type 5 hemagglutinin (H) protein and a type 1 neuraminidase (N) protein. There are 18 known types of hemagglutinin and 11 known types of neuraminidase, so, in theory, 198 different combinations of these proteins are possible.[5][6]
Some variants are identified and named according to the isolate they resemble, thus are presumed to share lineage (example Fujian flu virus-like); according to their typical host (example human flu virus); according to their subtype (example H3N2); and according to their deadliness (example LP, low pathogenic). So a flu from a virus similar to the isolate A/Fujian/411/2002(H3N2) is called Fujian flu, human flu, and H3N2 flu.
Variants are sometimes named according to the species (host) in which the strain is endemic or to which it is adapted. The main variants named using this convention are:
Variants have also sometimes been named according to their deadliness in poultry, especially chickens:
- Low pathogenic avian influenza (LPAI)
- Highly pathogenic avian influenza (HPAI), also called deadly flu or death flu
Most known strains are extinct strains. For example, the annual flu subtype H3N2 no longer contains the strain that caused the Hong Kong flu.
Annual flu
The annual flu (also called "seasonal flu" or "human flu") in the US. "results in approximately 36,000 deaths and more than 200,000 hospitalizations each year. In addition to this human toll, influenza is annually responsible for a total cost of over $10 billion in the U.S."[10] Globally the toll of influenza virus is estimated at 290,000–645,000 deaths annually, exceeding previous estimates.[11]
The annually updated, trivalent influenza vaccine consists of hemagglutinin (HA) surface glycoprotein components from influenza H3N2, H1N1, and B influenza viruses.[12]
Measured resistance to the standard antiviral drugs amantadine and rimantadine in H3N2 has increased from 1% in 1994 to 12% in 2003 to 91% in 2005.
"Contemporary human H3N2 influenza viruses are now endemic in pigs in southern China and can reassort with avian H5N1 viruses in this intermediate host."[13]
Structure and genetics
Influenza type A viruses are very similar in structure to influenza viruses types B, C, and D.[19] The virus particle (also called the virion) is 80–120 nanometers in diameter such that the smallest virions adopt an elliptical shape.[20][18] The length of each particle varies considerably, owing to the fact that influenza is pleomorphic, and can be in excess of many tens of micrometers, producing filamentous virions.[21] Confusion about the nature of influenza virus pleomorphy stems from the observation that lab adapted strains typically lose the ability to form filaments[22] and that these lab adapted strains were the first to be visualized by electron microscopy.[23] Despite these varied shapes, the virions of all influenza type A viruses are similar in composition. They are all made up of a viral envelope containing two main types of proteins, wrapped around a central core.[24]
The two large proteins found on the outside of viral particles are hemagglutinin (HA) and neuraminidase (NA). HA is a protein that mediates binding of the virion to target cells and entry of the viral genome into the target cell. NA is involved in release from the abundant non-productive attachment sites present in mucus[25] as well as the release of progeny virions from infected cells.[26] These proteins are usually the targets for antiviral drugs.[27] Furthermore, they are also the antigen proteins to which a host's antibodies can bind and trigger an immune response. Influenza type A viruses are categorized into subtypes based on the type of these two proteins on the surface of the viral envelope. There are 16 subtypes of HA and 9 subtypes of NA known, but only H 1, 2 and 3, and N 1 and 2 are commonly found in humans.[28]
The central core of a virion contains the viral genome and other viral proteins that package and protect the genetic material. Unlike the genomes of most organisms (including humans, animals, plants, and bacteria) which are made up of double-stranded DNA, many viral genomes are made up of a different, single-stranded nucleic acid called RNA. Unusually for a virus, though, the influenza type A virus genome is not a single piece of RNA; instead, it consists of segmented pieces of negative-sense RNA, each piece containing either one or two genes which code for a gene product (protein).[24] The term negative-sense RNA just implies that the RNA genome cannot be translated into protein directly; it must first be transcribed to positive-sense RNA before it can be translated into protein products. The segmented nature of the genome allows for the exchange of entire genes between different viral strains.[24]

The entire Influenza A virus genome is 13,588 bases long and is contained on eight RNA segments that code for at least 10 but up to 14 proteins, depending on the strain. The relevance or presence of alternate gene products can vary:[29]
- Segment 1 encodes RNA polymerase subunit (PB2).
- Segment 2 encodes RNA polymerase subunit (PB1) and the PB1-F2 protein, which induces cell death, by using different reading frames from the same RNA segment.
- Segment 3 encodes RNA polymerase subunit (PA) and the PA-X protein, which has a role in host transcription shutoff.[30]
- Segment 4 encodes for HA (hemagglutinin). About 500 molecules of hemagglutinin are needed to make one virion. HA determines the extent and severity of a viral infection in a host organism.
- Segment 5 encodes NP, which is a nucleoprotein.
- Segment 6 encodes NA (neuraminidase). About 100 molecules of neuraminidase are needed to make one virion.
- Segment 7 encodes two matrix proteins (M1 and M2) by using different reading frames from the same RNA segment. About 3,000 matrix protein molecules are needed to make one virion.
- Segment 8 encodes two distinct non-structural proteins (NS1 and NEP) by using different reading frames from the same RNA segment.
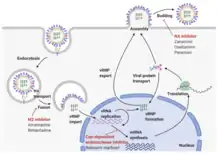
The RNA segments of the viral genome have complementary base sequences at the terminal ends, allowing them to bond to each other with hydrogen bonds.[26] Transcription of the viral (-) sense genome (vRNA) can only proceed after the PB2 protein binds to host capped RNAs, allowing for the PA subunit to cleave several nucleotides after the cap. This host-derived cap and accompanied nucleotides serve as the primer for viral transcription initiation. Transcription proceeds along the vRNA until a stretch of several uracil bases is reached, initiating a 'stuttering' whereby the nascent viral mRNA is poly-adenylated, producing a mature transcript for nuclear export and translation by host machinery.[31]
The RNA synthesis takes place in the cell nucleus, while the synthesis of proteins takes place in the cytoplasm. Once the viral proteins are assembled into virions, the assembled virions leave the nucleus and migrate towards the cell membrane.[32] The host cell membrane has patches of viral transmembrane proteins (HA, NA, and M2) and an underlying layer of the M1 protein which assist the assembled virions to budding through the membrane, releasing finished enveloped viruses into the extracellular fluid.[32]
Multiplicity reactivation
Influenza virus is able to undergo multiplicity reactivation after inactivation by UV radiation,[33][34] or by ionizing radiation.[35] If any of the eight RNA strands that make up the genome contains damage that prevents replication or expression of an essential gene, the virus is not viable when it alone infects a cell (a single infection). However, when two or more damaged viruses infect the same cell (multiple infection), viable progeny viruses can be produced provided each of the eight genomic segments is present in at least one undamaged copy. That is, multiplicity reactivation can occur.
Upon infection, influenza virus induces a host response involving increased production of reactive oxygen species, and this can damage the virus genome.[36] If, under natural conditions, virus survival is ordinarily vulnerable to the challenge of oxidative damage, then multiplicity reactivation is likely selectively advantageous as a kind of genomic repair process. It has been suggested that multiplicity reactivation involving segmented RNA genomes may be similar to the earliest evolved form of sexual interaction in the RNA world that likely preceded the DNA world.[37] (Also see RNA world hypothesis.)
Human influenza virus
"Human influenza virus" usually refers to those subtypes that spread widely among humans. H1N1, H1N2, and H3N2 are the only known influenza A virus subtypes currently circulating among humans.[38]
Genetic factors in distinguishing between "human flu viruses" and "avian influenza viruses" include:
- PB2: (RNA polymerase): Amino acid (or residue) position 627 in the PB2 protein encoded by the PB2 RNA gene. Until H5N1, all known avian influenza viruses had a Glu at position 627, while all human influenza viruses had a lysine.
- HA: (hemagglutinin): Avian influenza HA binds alpha 2–3 sialic acid receptors, while human influenza HA binds alpha 2–6 sialic acid receptors. Swine influenza viruses have the ability to bind both types of sialic acid receptors.
Human flu symptoms usually include fever, cough, sore throat, muscle aches, conjunctivitis and, in severe cases, breathing problems and pneumonia that may be fatal. The severity of the infection will depend in large part on the state of the infected person's immune system and if the victim has been exposed to the strain before, and is therefore partially immune. Follow-up studies on the impact of statins on influenza virus replication show that pre-treatment of cells with atorvastatin suppresses virus growth in culture.[39]
Highly pathogenic H5N1 avian influenza in a human is far worse, killing 50% of humans who catch it. In one case, a boy with H5N1 experienced diarrhea followed rapidly by a coma without developing respiratory or flu-like symptoms.[40]
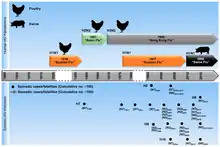
The influenza A virus subtypes that have been confirmed in humans, ordered by the number of known human pandemic deaths, are:
- H1N1 caused "Spanish flu" in 1918 and the 2009 swine flu pandemic
- H2N2 caused "Asian flu" in the late 1950s
- H3N2 caused "Hong Kong flu" in the late 1960s
- H5N1 considered a global influenza pandemic threat through its spread in the mid-2000s
- H7N9 is responsible for a 2013 epidemic in China[41] and considered by Dr. Michael Greger, author of How Not to Die, to have the greatest pandemic threat of the Influenza A viruses[42]
- H7N7 has some zoonotic potential: it has rarely caused disease in humans[43][44]
- H1N2 is currently endemic in pigs but not in humans.[45]
- H9N2, H7N2, H7N3, H5N2, and H10N7.
- H1N1
- H1N1 was responsible for the 2009 pandemic in both human and pig populations. A variant of H1N1 was responsible for the Spanish flu pandemic that killed some 50 million to 100 million people worldwide over about a year in 1918 and 1919.[46] Another variant was named a pandemic threat in the 2009 flu pandemic. Controversy arose in October 2005, after the H1N1 genome was published in the journal, Science, because of fears that this information could be used for bioterrorism.[47]
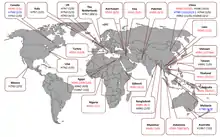 Human cases and fatalities caused by different influenza A virus subtypes
Human cases and fatalities caused by different influenza A virus subtypes
- H1N2
- H1N2 is currently endemic in both human and pig populations. The new H1N2 strain appears to have resulted from the reassortment of the genes of the currently circulating influenza H1N1 and H3N2 subtypes. The hemagglutinin protein of the H1N2 virus is similar to that of the currently circulating H1N1 viruses, and the neuraminidase protein is similar to that of the current H3N2 viruses.
- H2N2
- The Asian flu, a pandemic outbreak of H2N2 avian influenza, originated in China in 1957, spread worldwide that same year during which an influenza vaccine was developed, lasted until 1958 and caused between one and four million deaths.
- H3N2
- H3N2 is currently endemic in both human and pig populations. It evolved from H2N2 by antigenic shift and caused the Hong Kong flu pandemic of 1968, and 1969, that killed up to 750,000.[48] A severe form of the H3N2 virus killed several children in the United States in late 2003.[49]
- The dominant strain of annual flu in January 2006 was H3N2. Measured resistance to the standard antiviral drugs amantadine and rimantadine in H3N2 increased from 1% in 1994 to 12% in 2003 to 91% in 2005.[50] Human H3N2 influenza viruses are now endemic in pigs in southern China, where they circulate together with avian H5N1 viruses.[13]
- H5N1
 |
H5N1 is the world's major influenza pandemic threat.
- H5N2
- Japan's Health Ministry said January 2006 that poultry farm workers in Ibaraki prefecture may have been exposed to H5N2 in 2005.[51] The H5N2 antibody titers of paired sera of 13 subjects increased fourfold or more.[52]
- H5N9
- A highly pathogenic strain of H5N9 caused a minor flu outbreak in 1966 in Ontario and Manitoba, Canada in turkeys.[53]
- H7N2
- One person in New York in 2003, and one person in Virginia in 2002, were found to have serologic evidence of infection with H7N2. Both fully recovered.[54]
- H7N3
- In North America, the presence of avian influenza strain H7N3 was confirmed at several poultry farms in British Columbia in February 2004. As of April 2004, 18 farms had been quarantined to halt the spread of the virus. Two cases of humans with avian influenza have been confirmed in that region. "Symptoms included conjunctivitis and mild influenza-like illness."[55] Both fully recovered.
- H7N7
- H7N7 has unusual zoonotic potential. In 2003 in the Netherlands, 89 people were confirmed to have H7N7 influenza virus infection following an outbreak in poultry on several farms. One death was recorded.
- H7N9
- On 2 April 2013, the Centre for Health Protection (CHP) of the Department of Health of Hong Kong confirmed four more cases in Jiangsu province in addition to the three cases initially reported on 31 March 2013.[56] This virus also has the greatest potential for an influenza pandemic among all of the Influenza A subtypes.[57]
- H9N2
- Low pathogenic avian influenza A (H9N2) infection was confirmed in 1999, in China and Hong Kong in two children, and in 2003 in Hong Kong in one child. All three fully recovered.[54]
- H10N7
- In 2004, in Egypt, H10N7 was reported for the first time in humans. It caused illness in two infants in Egypt. One child’s father was a poultry merchant.[58]
Evolution
.jpg.webp)
According to Jeffery Taubenberger:
- "All influenza A pandemics since [the Spanish flu pandemic], and indeed almost all cases of influenza A worldwide (excepting human infections from avian viruses such as H5N1 and H7N7), have been caused by descendants of the 1918 virus, including "drifted" H1N1 viruses and reassorted H2N2 and H3N2 viruses. The latter are composed of key genes from the 1918 virus, updated by subsequently incorporated avian influenza genes that code for novel surface proteins, making the 1918 virus indeed the "mother" of all pandemics."[59]
Researchers from the National Institutes of Health used data from the Influenza Genome Sequencing Project and concluded that during the ten-year period examined, most of the time the hemagglutinin gene in H3N2 showed no significant excess of mutations in the antigenic regions while an increasing variety of strains accumulated. This resulted in one of the variants eventually achieving higher fitness, becoming dominant, and in a brief interval of rapid evolution, rapidly sweeping through the population and eliminating most other variants.[60]
In the short-term evolution of influenza A virus, a 2006 study found that stochastic, or random, processes are key factors.[61] Influenza A virus HA antigenic evolution appears to be characterized more by punctuated, sporadic jumps as opposed to a constant rate of antigenic change.[62] Using phylogenetic analysis of 413 complete genomes of human influenza A viruses that were collected throughout the state of New York, the authors of Nelson et al. 2006 were able to show that genetic diversity, and not antigenic drift, shaped the short-term evolution of influenza A via random migration and reassortment. The evolution of these viruses is dominated more by the random importation of genetically different viral strains from other geographic locations and less by natural selection. Within a given season, adaptive evolution is infrequent and had an overall weak effect as evidenced from the data gathered from the 413 genomes. Phylogenetic analysis revealed the different strains were derived from newly imported genetic material as opposed to isolates that had been circulating in New York in previous seasons. Therefore, the gene flow in and out of this population, and not natural selection, was more important in the short term.
Other animals
- See H5N1 for the current epizootic (an epidemic in nonhumans) and panzootic (a disease affecting animals of many species especially over a wide area) of H5N1 influenza
- Avian influenza
Fowl act as natural asymptomatic carriers of influenza A viruses. Prior to the current H5N1 epizootic, strains of influenza A virus had been demonstrated to be transmitted from wildfowl to only birds, pigs, horses, seals, whales and humans; and only between humans and pigs and between humans and domestic fowl; and not other pathways such as domestic fowl to horse.[63]
Wild aquatic birds are the natural hosts for a large variety of influenza A viruses. Occasionally, viruses are transmitted from these birds to other species and may then cause devastating outbreaks in domestic poultry or give rise to human influenza pandemics.[3][4]
H5N1 has been shown to be transmitted to tigers, leopards, and domestic cats that were fed uncooked domestic fowl (chickens) with the virus. H3N8 viruses from horses have crossed over and caused outbreaks in dogs. Laboratory mice have been infected successfully with a variety of avian flu genotypes.[64]
Influenza A viruses spread in the air and in manure, and survives longer in cold weather. They can also be transmitted by contaminated feed, water, equipment, and clothing; however, there is no evidence the virus can survive in well-cooked meat. Symptoms in animals vary, but virulent strains can cause death within a few days. Avian influenza viruses that the World Organisation for Animal Health and others test for to control poultry disease include H5N1, H7N2, H1N7, H7N3, H13N6, H5N9, H11N6, H3N8, H9N2, H5N2, H4N8, H10N7, H2N2, H8N4, H14N5, H6N5, and H12N5.
- Known outbreaks of highly pathogenic flu in poultry 1959–2003[65]
| Year | Area | Affected | Subtype |
|---|---|---|---|
| 1959 | Scotland | Chicken | H5N1 |
| 1963 | England | Turkey | H7N3 |
| 1966 | Ontario (Canada) | Turkey | H5N9 |
| 1976 | Victoria (Australia) | Chicken | H7N7 |
| 1979 | Germany | Chicken | H7N7 |
| 1979 | England | Turkey | H7N7 |
| 1983 | Pennsylvania (US)* | Chicken, turkey | H5N2 |
| 1983 | Ireland | Turkey | H5N8 |
| 1985 | Victoria (Australia) | Chicken | H7N7 |
| 1991 | England | Turkey | H5N1 |
| 1992 | Victoria (Australia) | Chicken | H7N3 |
| 1994 | Queensland (Australia) | Chicken | H7N3 |
| 1994 | Mexico* | Chicken | H5N2 |
| 1994 | Pakistan* | Chicken | H7N3 |
| 1997 | New South Wales (Australia) | Chicken | H7N4 |
| 1997 | Hong Kong (China)* | Chicken | H5N1 |
| 1997 | Italy | Chicken | H5N2 |
| 1999 | Italy* | Turkey | H7N1 |
| 2002 | Hong Kong (China) | Chicken | H5N1 |
| 2002 | Chile | Chicken | H7N3 |
| 2003 | Netherlands* | Chicken | H7N7 |
*Outbreaks with significant spread to numerous farms, resulting in great economic losses. Most other outbreaks involved little or no spread from the initially infected farms.
More than 400 harbor seal deaths were recorded in New England between December 1979 and October 1980, from acute pneumonia caused by the influenza virus, A/Seal/Mass/1/180 (H7N7).[66]
- Swine flu
- Swine influenza (or "pig influenza") refers to a subset of Orthomyxoviridae that create influenza and are endemic in pigs. The species of Orthomyxoviridae that can cause flu in pigs are influenza A virus and influenza C virus, but not all genotypes of these two species infect pigs. The known subtypes of influenza A virus that create influenza and are endemic in pigs are H1N1, H1N2, H3N1 and H3N2. In 1997, H3N2 viruses from humans entered the pig population, causing widespread disease among pigs.[67]
- Horse flu
- Horse flu (or "equine influenza") refers to varieties of influenza A virus that affect horses. Horse flu viruses were only isolated in 1956. The two main types of virus are called equine-1 (H7N7), which commonly affects horse heart muscle, and equine-2 (H3N8), which is usually more severe. H3N8 viruses from horses have infected dogs.[67]
- Dog flu
- Dog flu (or "canine influenza") refers to varieties of influenza A virus that affect dogs. The equine influenza virus H3N8 was found to infect and kill – with respiratory illness – greyhound race dogs at a Florida racetrack in January 2004.
- Bat flu
- Bat flu (or "Bat influenza") refers to the H17N10 and H18N11 influenza A virus strains that were discovered in Central and South American fruit bats as well as a H9N2 virus isolated from the Egyptian fruit bat.[68] Until now it is unclear whether these bat-derived viruses are circulating in any non-bat species and whether they pose a zoonotic threat. Initial characterization of the H18N11 subtype, however, suggests that this bat influenza virus is not well adapted to any other species than bats.[69]
- H3N8
- H3N8 is now endemic in birds, horses and dogs.
Subtype list
Influenza A virus has the following subtypes:
- Influenza A virus subtype H1N1
- Influenza A virus subtype H1N2
- Influenza A virus subtype H2N2
- Influenza A virus subtype H2N3
- Influenza A virus subtype H3N1
- Influenza A virus subtype H3N2
- Influenza A virus subtype H3N8
- Influenza A virus subtype H5N1
- Influenza A virus subtype H5N2
- Influenza A virus subtype H5N3
- Influenza A virus subtype H5N6
- Influenza A virus subtype H5N8
- Influenza A virus subtype H5N9
- Influenza A virus subtype H6N1
- Influenza A virus subtype H6N2
- Influenza A virus subtype H7N1
- Influenza A virus subtype H7N2
- Influenza A virus subtype H7N3
- Influenza A virus subtype H7N4
- Influenza A virus subtype H7N7
- Influenza A virus subtype H7N9
- Influenza A virus subtype H9N2
- Influenza A virus subtype H10N7
- Influenza A virus subtype H10N8
- Influenza A virus subtype H11N2
- Influenza A virus subtype H11N9
- Influenza A virus subtype H17N10
- Influenza A virus subtype H18N11
Notes
- "Taxonomy". International Committee on Taxonomy of Viruses (ICTV). Retrieved 19 July 2018.
- "Avian influenza (" bird flu") – Fact sheet". WHO.
- Klenk H, Matrosovich M, Stech J (2008). "Avian Influenza: Molecular Mechanisms of Pathogenesis and Host Range". In Mettenleiter TC, Sobrino F (eds.). Animal Viruses: Molecular Biology. Caister Academic Press. ISBN 978-1-904455-22-6.
- Kawaoka Y, ed. (2006). Influenza Virology: Current Topics. Caister Academic Press. ISBN 978-1-904455-06-6.
- "Influenza Type A Viruses and Subtypes". Centers for Disease Control and Prevention. 2 April 2013. Retrieved 13 June 2013.
- Tong S, Zhu X, Li Y, Shi M, Zhang J, Bourgeois M, Yang H, Chen X, Recuenco S, Gomez J, Chen LM, Johnson A, Tao Y, Dreyfus C, Yu W, McBride R, Carney PJ, Gilbert AT, Chang J, Guo Z, Davis CT, Paulson JC, Stevens J, Rupprecht CE, Holmes EC, Wilson IA, Donis RO (October 2013). "New world bats harbor diverse influenza A viruses". PLOS Pathogens. 9 (10): e1003657. doi:10.1371/journal.ppat.1003657. PMC 3794996. PMID 24130481.
- "Unique new flu virus found in bats". NHS Choices. 1 March 2012. Retrieved 16 May 2012.
- Tong S, Li Y, Rivailler P, Conrardy C, Castillo DA, Chen LM, Recuenco S, Ellison JA, Davis CT, York IA, Turmelle AS, Moran D, Rogers S, Shi M, Tao Y, Weil MR, Tang K, Rowe LA, Sammons S, Xu X, Frace M, Lindblade KA, Cox NJ, Anderson LJ, Rupprecht CE, Donis RO (March 2012). "A distinct lineage of influenza A virus from bats". Proceedings of the National Academy of Sciences of the United States of America. 109 (11): 4269–74. Bibcode:2012PNAS..109.4269T. doi:10.1073/pnas.1116200109. PMC 3306675. PMID 22371588.
- Gallagher J (29 July 2011). "'Super antibody' fights off flu". BBC News. Retrieved 29 July 2011.
- whitehouse.gov Archived 9 January 2009 at the Wayback Machine National Strategy for Pandemic Influenza – Introduction – "Although remarkable advances have been made in science and medicine during the past century, we are constantly reminded that we live in a universe of microbes – viruses, bacteria, protozoa and fungi that are forever changing and adapting themselves to the human host and the defenses that humans create. Influenza viruses are notable for their resilience and adaptability. While science has been able to develop highly effective vaccines and treatments for many infectious diseases that threaten public health, acquiring these tools is an ongoing challenge with the influenza virus. Changes in the genetic makeup of the virus require us to develop new vaccines on an annual basis and forecast which strains are likely to predominate. As a result, and despite annual vaccinations, the US faces a burden of influenza that results in approximately 36,000 deaths and more than 200,000 hospitalizations each year. In addition to this human toll, influenza is annually responsible for a total cost of over $10 billion in the US. A pandemic, or worldwide outbreak of a new influenza virus, could dwarf this impact by overwhelming our health and medical capabilities, potentially resulting in hundreds of thousands of deaths, millions of hospitalizations, and hundreds of billions of dollars in direct and indirect costs. This Strategy will guide our preparedness and response activities to mitigate that impact."
- Iuliano AD, Roguski KM, Chang HH, Muscatello DJ, Palekar R, Tempia S, Cohen C, Gran JM, Schanzer D, Cowling BJ, Wu P, Kyncl J, Ang LW, Park M, Redlberger-Fritz M, Yu H, Espenhain L, Krishnan A, Emukule G, van Asten L, Pereira da Silva S, Aungkulanon S, Buchholz U, Widdowson MA, Bresee JS (March 2018). "Estimates of global seasonal influenza-associated respiratory mortality: a modelling study". Lancet. 391 (10127): 1285–1300. doi:10.1016/s0140-6736(17)33293-2. PMC 5935243. PMID 29248255.
- Daum LT, Shaw MW, Klimov AI, Canas LC, Macias EA, Niemeyer D, Chambers JP, Renthal R, Shrestha SK, Acharya RP, Huzdar SP, Rimal N, Myint KS, Gould P (August 2005). "Influenza A (H3N2) outbreak, Nepal". Emerging Infectious Diseases. 11 (8): 1186–91. doi:10.3201/eid1108.050302. PMC 3320503. PMID 16102305.
"The 2003–2004 influenza season was severe in terms of its impact on illness because of widespread circulation of antigenically distinct influenza A (H3N2) Fujian-like viruses. These viruses first appeared late during the 2002–2003 influenza season and continued to persist as the dominant circulating strain throughout the subsequent 2003–2004 influenza season, replacing the A/Panama/2007/99-like H3N2 viruses (1). Of the 172 H3N2 viruses genetically characterized by the Department of Defense in 2003–2004, only one isolate (from Thailand) belonged to the A/Panama-like lineage. In February 2003, the World Health Organization (WHO) changed the H3N2 component for the 2004–2005 influenza vaccine to afford protection against the widespread emergence of Fujian-like viruses (2). The annually updated trivalent vaccine consists of hemagglutinin (HA) surface glycoprotein components from influenza H3N2, H1N1, and B viruses." - Mahmoud 2005, p. 126
"H5N1 virus is now endemic in poultry in Asia (Table 2-1) and has gained an entrenched ecological niche from which to present a long-term pandemic threat to humans. At present, these viruses are poorly transmitted from poultry to humans, and there is no conclusive evidence of human-to-human transmission. However, continued, extensive exposure of the human population to H5N1 viruses increases the likelihood that the viruses will acquire the necessary characteristics for efficient human-to-human transmission through genetic mutation or reassortment with a prevailing human influenza A virus. Furthermore, contemporary human H3N2 influenza viruses are now endemic in pigs in southern China (Peiris et al., 2001) and can reassort with avian H5N1 viruses in this 'intermediate host.' Therefore, it is imperative that outbreaks of H5N1 disease in poultry in Asia are rapidly and sustainably controlled. The seasonality of the disease in poultry, together with the control measures already implemented, are likely to reduce temporarily the frequency of H5N1 influenza outbreaks and the probability of human infection." - Gallagher J (29 July 2011). "'Super antibody' fights off flu". BBC News.
- "Scientists hail the prospect of a universal vaccine for flu". 29 July 2011.
- Chan AL (28 July 2011). "Universal Flu Vaccine on the Horizon: Researchers Find 'Super Antibody'". HuffPost.
- "Details – Public Health Image Library(PHIL)". phil.cdc.gov. Retrieved 24 April 2018.
- Sugita Y, Noda T, Sagara H, Kawaoka Y (November 2011). "Ultracentrifugation deforms unfixed influenza A virions". The Journal of General Virology. 92 (Pt 11): 2485–93. doi:10.1099/vir.0.036715-0. PMC 3352361. PMID 21795472.
- Nakatsu S, Murakami S, Shindo K, Horimoto T, Sagara H, Noda T, Kawaoka Y (March 2018). "Influenza C and D Viruses Package Eight Organized Ribonucleoprotein Complexes". Journal of Virology. 92 (6): e02084–17. doi:10.1128/jvi.02084-17. PMC 5827381. PMID 29321324.
- Noda T (2011). "Native morphology of influenza virions". Frontiers in Microbiology. 2: 269. doi:10.3389/fmicb.2011.00269. PMC 3249889. PMID 22291683.
- Dadonaite B, Vijayakrishnan S, Fodor E, Bhella D, Hutchinson EC (August 2016). "Filamentous influenza viruses". The Journal of General Virology. 97 (8): 1755–64. doi:10.1099/jgv.0.000535. PMC 5935222. PMID 27365089.
- Seladi-Schulman J, Steel J, Lowen AC (December 2013). "Spherical influenza viruses have a fitness advantage in embryonated eggs, while filament-producing strains are selected in vivo". Journal of Virology. 87 (24): 13343–53. doi:10.1128/JVI.02004-13. PMC 3838284. PMID 24089563.
- Mosley VM, Wyckoff RW (March 1946). "Electron micrography of the virus of influenza". Nature. 157 (3983): 263. Bibcode:1946Natur.157..263M. doi:10.1038/157263a0. PMID 21016866. S2CID 6478026.
- Bouvier NM, Palese P (September 2008). "The biology of influenza viruses". Vaccine. 26 (Suppl 4): D49–53. doi:10.1016/j.vaccine.2008.07.039. PMC 3074182. PMID 19230160.
- Cohen M, Zhang XQ, Senaati HP, Chen HW, Varki NM, Schooley RT, Gagneux P (November 2013). "Influenza A penetrates host mucus by cleaving sialic acids with neuraminidase". Virology Journal. 10: 321. doi:10.1186/1743-422x-10-321. PMC 3842836. PMID 24261589.
- Suzuki Y (March 2005). "Sialobiology of influenza: molecular mechanism of host range variation of influenza viruses". Biological & Pharmaceutical Bulletin. 28 (3): 399–408. doi:10.1248/bpb.28.399. PMID 15744059.
- Wilson JC, von Itzstein M (July 2003). "Recent strategies in the search for new anti-influenza therapies". Current Drug Targets. 4 (5): 389–408. doi:10.2174/1389450033491019. PMID 12816348.
- Lynch JP, Walsh EE (April 2007). "Influenza: evolving strategies in treatment and prevention". Seminars in Respiratory and Critical Care Medicine. 28 (2): 144–58. doi:10.1055/s-2007-976487. PMID 17458769.
- Eisfeld AJ, Neumann G, Kawaoka Y (January 2015). "At the centre: influenza A virus ribonucleoproteins". Nature Reviews. Microbiology. 13 (1): 28–41. doi:10.1038/nrmicro3367. PMC 5619696. PMID 25417656.
- Khaperskyy DA, Schmaling S, Larkins-Ford J, McCormick C, Gaglia MM (February 2016). "Selective Degradation of Host RNA Polymerase II Transcripts by Influenza A Virus PA-X Host Shutoff Protein". PLOS Pathogens. 12 (2): e1005427. doi:10.1371/journal.ppat.1005427. PMC 4744033. PMID 26849127.
- Te Velthuis AJ, Fodor E (August 2016). "Influenza virus RNA polymerase: insights into the mechanisms of viral RNA synthesis". Nature Reviews. Microbiology. 14 (8): 479–93. doi:10.1038/nrmicro.2016.87. PMC 4966622. PMID 27396566.
- Smith AE, Helenius A (April 2004). "How viruses enter animal cells". Science. 304 (5668): 237–42. Bibcode:2004Sci...304..237S. doi:10.1126/science.1094823. PMID 15073366. S2CID 43062708.
- Barry RD (August 1961). "The multiplication of influenza virus. II. Multiplicity reactivation of ultraviolet irradiated virus". Virology. 14 (4): 398–405. doi:10.1016/0042-6822(61)90330-0. hdl:1885/109240. PMID 13687359.
- Henle W, Liu OC (October 1951). "Studies on host-virus interactions in the chick embryo-influenza virus system. VI. Evidence for multiplicity reactivation of inactivated virus". The Journal of Experimental Medicine. 94 (4): 305–22. doi:10.1084/jem.94.4.305. PMC 2136114. PMID 14888814.
- Gilker JC, Pavilanis V, Ghys R (June 1967). "Multiplicity reactivation in gamma irradiated influenza viruses". Nature. 214 (5094): 1235–7. Bibcode:1967Natur.214.1235G. doi:10.1038/2141235a0. PMID 6066111. S2CID 4200194.
- Peterhans E (May 1997). "Oxidants and antioxidants in viral diseases: disease mechanisms and metabolic regulation". The Journal of Nutrition. 127 (5 Suppl): 962S–965S. doi:10.1093/jn/127.5.962S. PMID 9164274.
- Bernstein H, Byerly HC, Hopf FA, Michod RE (October 1984). "Origin of sex". Journal of Theoretical Biology. 110 (3): 323–51. doi:10.1016/S0022-5193(84)80178-2. PMID 6209512.
- CDC Key Facts About Avian Influenza (Bird Flu) and Avian Influenza A (H5N1) Virus
- Episcopio D, Aminov S, Benjamin S, Germain G, Datan E, Landazuri J, Lockshin RA, Zakeri Z (April 2019). "Atorvastatin restricts the ability of influenza virus to generate lipid droplets and severely suppresses the replication of the virus". The FASEB Journal. 33 (8): 9516–9525. doi:10.1096/fj.201900428RR. PMC 6662987. PMID 31125254.
- de Jong MD, Bach VC, Phan TQ, Vo MH, Tran TT, Nguyen BH, Beld M, Le TP, Truong HK, Nguyen VV, Tran TH, Do QH, Farrar J (February 2005). "Fatal avian influenza A (H5N1) in a child presenting with diarrhea followed by coma". The New England Journal of Medicine. 352 (7): 686–91. doi:10.1056/NEJMoa044307. PMID 15716562. S2CID 17703507.
- "New bird flu strain in China "one of the most lethal" warns WHO". MercoPress. Retrieved 6 December 2020.
- "The next pandemic and how to head it off: eat a plant-based diet". South China Morning Post. 19 October 2020. Retrieved 6 December 2020.
- Boston, 677 Huntington Avenue; Ma 02115 +1495‑1000 (24 October 2013). "Making the leap". News. Retrieved 6 December 2020.
- Ungchusak, Kumnuan; Auewarakul, Prasert; Dowell, Scott F.; Kitphati, Rungrueng; Auwanit, Wattana; Puthavathana, Pilaipan; Uiprasertkul, Mongkol; Boonnak, Kobporn; Pittayawonganon, Chakrarat; Cox, Nancy J.; Zaki, Sherif R. (27 January 2005). "Probable Person-to-Person Transmission of Avian Influenza A (H5N1)". New England Journal of Medicine. 352 (4): 333–340. doi:10.1056/NEJMoa044021. ISSN 0028-4793. PMID 15668219.
- "First Human Case Of Rare H1N2 Flu Variant Reported In Canada". IFLScience. Retrieved 6 December 2020.
- Mahmoud 2005, p. 7
- "NOVA | scienceNOW | Reviving the Virus (non-Flash) | PBS". www.pbs.org. Retrieved 6 December 2020.
- Detailed chart of its evolution here Archived 9 May 2009 at the Wayback Machine at PDF called Ecology and Evolution of the Flu
- Mahmoud 2005, p. 115
"There is particular pressure to recognize and heed the lessons of past influenza pandemics in the shadow of the worrisome 2003–2004 flu season. An early-onset, severe form of influenza A H3N2 made headlines when it claimed the lives of several children in the United States in late 2003. As a result, stronger than usual demand for annual flu inactivated vaccine outstripped the vaccine supply, of which 10 to 20 percent typically goes unused. Because statistics on pediatric flu deaths had not been collected previously, it is unknown if the 2003–2004 season witnessed a significant change in mortality patterns." - Reason Archived 26 October 2006 at the Wayback Machine Altman LK (15 January 2006). "This Season's Flu Virus Is Resistant to 2 Standard Drugs". The New York Times.
- CBS News article Dozens in Japan May Have Mild Bird Flu January 2006.
- Ogata T, Yamazaki Y, Okabe N, Nakamura Y, Tashiro M, Nagata N, Itamura S, Yasui Y, Nakashima K, Doi M, Izumi Y, Fujieda T, Yamato S, Kawada Y (July 2008). "Human H5N2 avian influenza infection in Japan and the factors associated with high H5N2-neutralizing antibody titer". Journal of Epidemiology. 18 (4): 160–6. doi:10.2188/jea.JE2007446. PMC 4771585. PMID 18603824.
- WHO
- CDC Avian Influenza Infection in Humans
- Tweed SA, Skowronski DM, David ST, Larder A, Petric M, Lees W, Li Y, Katz J, Krajden M, Tellier R, Halpert C, Hirst M, Astell C, Lawrence D, Mak A (December 2004). "Human illness from avian influenza H7N3, British Columbia". Emerging Infectious Diseases. 10 (12): 2196–9. doi:10.3201/eid1012.040961. PMC 3323407. PMID 15663860.
- Schnirring L (2 April 2013). "China reports 4 more H7N9 infections". CIDRAP News.
- "Avian Influenza A (H7N9) Virus | Avian Influenza (Flu)". www.cdc.gov. Retrieved 24 February 2017.
- niaid.nih.gov Archived 26 December 2005 at the Wayback Machine Timeline of Human Flu Pandemics
- Taubenberger JK, Morens DM (January 2006). "1918 Influenza: the mother of all pandemics". Emerging Infectious Diseases. 12 (1): 15–22. doi:10.3201/eid1201.050979. PMC 3291398. PMID 16494711.
- Science Daily article New Study Has Important Implications For Flu Surveillance published 27 October 2006
- Nelson MI, Simonsen L, Viboud C, Miller MA, Taylor J, George KS, Griesemer SB, Ghedin E, Ghedi E, Sengamalay NA, Spiro DJ, Volkov I, Grenfell BT, Lipman DJ, Taubenberger JK, Holmes EC (December 2006). "Stochastic processes are key determinants of short-term evolution in influenza a virus". PLOS Pathogens. 2 (12): e125. doi:10.1371/journal.ppat.0020125. PMC 1665651. PMID 17140286.
- Smith DJ, Lapedes AS, de Jong JC, Bestebroer TM, Rimmelzwaan GF, Osterhaus AD, Fouchier RA (July 2004). "Mapping the antigenic and genetic evolution of influenza virus". Science. 305 (5682): 371–6. Bibcode:2004Sci...305..371S. doi:10.1126/science.1097211. PMID 15218094. S2CID 1258353.
- Mahmoud 2005, p. 30
- Mahmoud 2005, p. 82
"Interestingly, recombinant influenza viruses containing the 1918 HA and NA and up to three additional genes derived from the 1918 virus (the other genes being derived from the A/WSN/33 virus) were all highly virulent in mice (Tumpey et al., 2004). Furthermore, expression microarray analysis performed on whole lung tissue of mice infected with the 1918 HA/ NA recombinant showed increased upregulation of genes involved in apoptosis, tissue injury, and oxidative damage (Kash et al., 2004). These findings were unusual because the viruses with the 1918 genes had not been adapted to mice. The completion of the sequence of the entire genome of the 1918 virus and the reconstruction and characterization of viruses with 1918 genes under appropriate biosafety conditions will shed more light on these findings and should allow a definitive examination of this explanation. Antigenic analysis of recombinant viruses possessing the 1918 HA and NA by hemagglutination inhibition tests using ferret and chicken antisera suggested a close relationship with the A/swine/Iowa/30 virus and H1N1 viruses isolated in the 1930s (Tumpey et al., 2004), further supporting data of Shope from the 1930s (Shope, 1936). Interestingly, when mice were immunized with different H1N1 virus strains, challenge studies using the 1918-like viruses revealed partial protection by this treatment, suggesting that current vaccination strategies are adequate against a 1918-like virus (Tumpey et al., 2004)." - "Avian influenza A(H5N1)- update 31: Situation (poultry) in Asia: need for a long-term response, comparison with previous outbreaks". Epidemic and Pandemic Alert and Response (EPR). WHO. 2004.
Known outbreaks of highly pathogenic flu in poultry 1959–2003. - Geraci JR, St Aubin DJ, Barker IK, Webster RG, Hinshaw VS, Bean WJ, Ruhnke HL, Prescott JH, Early G, Baker AS, Madoff S, Schooley RT (February 1982). "Mass mortality of harbor seals: pneumonia associated with influenza A virus". Science. 215 (4536): 1129–31. Bibcode:1982Sci...215.1129G. doi:10.1126/science.7063847. PMID 7063847.
More than 400 harbor seals, most of them immature, died along the New England coast between December 1979 and October 1980 of acute pneumonia associated with influenza virus, A/Seal/Mass/1/180 (H7N7). The virus has avian characteristics, replicates principally in mammals, and causes mild respiratory disease in experimentally infected seals. Concurrent infection with a previously undescribed mycoplasma or adverse environmental conditions may have triggered the epizootic. The similarities between this epizootic and other seal mortalities in the past suggest that these events may be linked by common biological and environmental factors.
- CDC Centers for Disease Control and Prevention – Transmission of Influenza A Viruses Between Animals and People
- Kandeil A, Gomaa MR, Shehata MM, El Taweel AN, Mahmoud SH, Bagato O (January 2019). "Isolation and Characterization of a Distinct Influenza A Virus from Egyptian Bats". Journal of Virology. 93 (2): e01059-18. doi:10.1128/JVI.01059-18. PMC 6321940. PMID 30381492.
- Ciminski K, Ran W, Gorka M, Lee J, Schinköthe J, Eckley M, Murrieta MA, Aboellail TA, Campbell CL, Ebel GD, Ma J, Pohlmann A, Franzke K, Ulrich R, Hoffmann D, Garcia-Sastre A, Ma W, Schountz T, Beer M, Schwemmle M (2019). "Bat influenza viruses transmit among bats but are poorly adapted to non-bat species". Nature Microbiology. 4 (12): 2298–2309. doi:10.1038/s41564-019-0556-9. PMC 7758811. PMID 31527796. S2CID 202580293.
Further reading
- Official sources
- Avian influenza and Influenza Pandemics from the Centers for Disease Control and Prevention
- Avian influenza FAQ from the World Health Organization
- Avian influenza information from the Food and Agriculture Organization
- U.S. Government's avian influenza information website
- European Centre for Disease Prevention and Control (ECDC) Stockholm, Sweden
- General information
- "The Bird Flu and You" Full-color poster provided by the Center for Technology and National Security Policy at the National Defense University, in collaboration with the National Security Health Policy Center
- Special issue on avian flu from Nature
- Nature Reports: Homepage: Avian Flu
- Beigel JH, Farrar J, Han AM, Hayden FG, Hyer R, de Jong MD, Lochindarat S, Nguyen TK, Nguyen TH, Tran TH, Nicoll A, Touch S, Yuen KY (September 2005). "Avian influenza A (H5N1) infection in humans". The New England Journal of Medicine. 353 (13): 1374–85. CiteSeerX 10.1.1.730.7890. doi:10.1056/NEJMra052211. PMID 16192482.
- Pandemic Influenza: Domestic Preparedness Efforts Congressional Research Service Report on Pandemic Preparedness.
- A guide to bird flu and its symptoms from BBC Health
- Mahmoud (2005). Stacey L. Knobler; Alison Mack; Mahmoud, Adel; Stanley M. Lemon (eds.). The threat of pandemic influenza : are we ready? : workshop summary / prepared for Forum on Microbial Threats, Board on Global Health. The National Academies Press. p. 285. ISBN 0-309-09504-2.
Highly pathogenic avian influenza virus is on every top ten list available for potential agricultural bioweapon agents
- Mahmoud AA, Institute of Medicine, Knobler S, Mack A (2005). The Threat of Pandemic Influenza: Are We Ready?: Workshop Summary. Washington, D.C: National Academies Press. ISBN 978-0-309-09504-4.
- Links to Bird Flu pictures (Hardin MD/Univ of Iowa)
- Kawaoka Y (2006). Influenza Virology: Current Topics. Caister Academic Pr. ISBN 978-1-904455-06-6.
- Sobrino F, Mettenleiter T (2008). Animal Viruses: Molecular Biology. Caister Academic Pr. ISBN 978-1-904455-22-6.
External links
- Influenza Research Database – Database of influenza genomic sequences and related information.
- Health-EU portal European Union response to influenza