Vector (molecular biology)
In molecular cloning, a vector is a DNA molecule used as a vehicle to artificially carry foreign genetic material into another cell, where it can be replicated and/or expressed (e.g., plasmid, cosmid, Lambda phages). A vector containing foreign DNA is termed recombinant DNA. The four major types of vectors are plasmids, viral vectors, cosmids, and artificial chromosomes. Of these, the most commonly used vectors are plasmids.[1] Common to all engineered vectors have an origin of replication, a multicloning site, and a selectable marker.
The vector itself is generally a DNA sequence that consists of an insert (transgene) and a larger sequence that serves as the "backbone" of the vector. The purpose of a vector which transfers genetic information to another cell is typically to isolate, multiply, or express the insert in the target cell. All vectors may be used for cloning and are therefore cloning vectors, but there are also vectors designed specially for cloning, while others may be designed specifically for other purposes, such as transcription and protein expression. Vectors designed specifically for the expression of the transgene in the target cell are called expression vectors, and generally have a promoter sequence that drives expression of the transgene. Simpler vectors called transcription vectors are only capable of being transcribed but not translated: they can be replicated in a target cell but not expressed, unlike expression vectors. Transcription vectors are used to amplify their insert.
The manipulation of DNA is normally conducted on E. coli vectors, which contain elements necessary for their maintenance in E. coli. However, vectors may also have elements that allow them to be maintained in another organism such as yeast, plant or mammalian cells, and these vectors are called shuttle vectors. Such vectors have bacterial or viral elements which may be transferred to the non-bacterial host organism, however other vectors termed intragenic vectors have also been developed to avoid the transfer of any genetic material from an alien species.[2]
Insertion of a vector into the target cell is usually called transformation for bacterial cells,[3] transfection for eukaryotic cells,[4] although insertion of a viral vector is often called transduction.[5]
Characteristics
Plasmids
Plasmids are double-stranded extra chromosomal and generally circular DNA sequences that are capable of replication using the host cell's replication machinery.[6] Plasmid vectors minimalistically consist of an origin of replication that allows for semi-independent replication of the plasmid in the host. Plasmids are found widely in many bacteria, for example in Escherichia coli, but may also be found in a few eukaryotes, for example in yeast such as Saccharomyces cerevisiae.[7] Bacterial plasmids may be conjugative/transmissible and non-conjugative:
- conjugative - mediate DNA transfer through conjugation and therefore spread rapidly among the bacterial cells of a population; e.g., F plasmid, many R and some col plasmids.
- nonconjugative - do not mediate DNA through conjugation, e.g., many R and col plasmids.
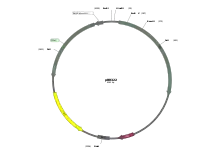
Plasmids with specially-constructed features are commonly used in laboratory for cloning purposes. These plasmid are generally non-conjugative but may have many more features, notably a "multiple cloning site" where multiple restriction enzyme cleavage sites allow for the insertion of a transgene insert. The bacteria containing the plasmids can generate millions of copies of the vector within the bacteria in hours, and the amplified vectors can be extracted from the bacteria for further manipulation. Plasmids may be used specifically as transcription vectors and such plasmids may lack crucial sequences for protein expression. Plasmids used for protein expression, called expression vectors, would include elements for translation of protein, such as a ribosome binding site, start and stop codons.
Viral vectors
Viral vectors are generally genetically engineered viruses carrying modified viral DNA or RNA that has been rendered noninfectious, but still contain viral promoters and also the transgene, thus allowing for translation of the transgene through a viral promoter. However, because viral vectors frequently are lacking infectious sequences, they require helper viruses or packaging lines for large-scale transfection. Viral vectors are often designed for permanent incorporation of the insert into the host genome, and thus leave distinct genetic markers in the host genome after incorporating the transgene. For example, retroviruses leave a characteristic retroviral integration pattern after insertion that is detectable and indicates that the viral vector has incorporated into the host genome.
Artificial chromosomes
Artificial chromosomes are manufactured chromosomes in the context of yeast artificial chromosomes (YACs), bacterial artificial chromosomes (BACs), or human artificial chromosomes (HACs). An artificial chromosome can carry a much larger DNA fragment than other vectors.[8] YACs and BACs can carry a DNA fragment up to 300,000 nucleotides long. Three structural necessities of an artificial chromosome include an origin of replication, a centromere, and telomeric end sequences.[9]
Transcription
Transcription of the cloned gene is a necessary component of the vector when expression of the gene is required: one gene may be amplified through transcription to generate multiple copies of mRNAs, the template on which protein may be produced through translation.[10] A larger number of mRNAs would express a greater amount of protein, and how many copies of mRNA are generated depends on the promoter used in the vector.[11] The expression may be constitutive, meaning that the protein is produced constantly in the background, or it may be inducible whereby the protein is expressed only under certain condition, for example when a chemical inducer is added. These two different types of expression depend on the types of promoter and operator used.
Viral promoters are often used for constitutive expression in plasmids and in viral vectors because they normally force constant transcription in many cell lines and types reliably.[12] Inducible expression depends on promoters that respond to the induction conditions: for example, the murine mammary tumor virus promoter only initiates transcription after dexamethasone application and the Drosophilia heat shock promoter only initiates after high temperatures.
Some vectors are designed for transcription only, for example for in vitro mRNA production. These vectors are called transcription vectors. They may lack the sequences necessary for polyadenylation and termination, therefore may not be used for protein production.
Expression
Expression vectors produce proteins through the transcription of the vector's insert followed by translation of the mRNA produced, they therefore require more components than the simpler transcription-only vectors. Expression in different host organism would require different elements, although they share similar requirements, for example a promoter for initiation of transcription, a ribosomal binding site for translation initiation, and termination signals.
Prokaryotes expression vector
- Promoter - commonly used inducible promoters are promoters derived from lac operon and the T7 promoter. Other strong promoters used include Trp promoter and Tac-Promoter, which are a hybrid of both the Trp and Lac Operon promoters.
- Ribosome binding site (RBS) - follows the promoter, and promotes efficient translation of the protein of interest.
- Translation initiation site - Shine-Dalgarno sequence enclosed in the RBS, 8 base-pairs upstream of the AUG start codon.
Eukaryotes expression vector
Eukaryote expression vectors require sequences that encode for:
- Polyadenylation tail: Creates a polyadenylation tail at the end of the transcribed pre-mRNA that protects the mRNA from exonucleases and ensures transcriptional and translational termination: stabilizes mRNA production.
- Minimal UTR length: UTRs contain specific characteristics that may impede transcription or translation, and thus the shortest UTRs or none at all are encoded for in optimal expression vectors.
- Kozak sequence: Vectors should encode for a Kozak sequence in the mRNA, which assembles the ribosome for translation of the mRNA.
Features
Modern artificially-constructed vectors contain essential components found in all vectors, and may contain other additional features found only in some vectors:
- Origin of replication: Necessary for the replication and maintenance of the vector in the host cell.
- Promoter: Promoters are used to drive the transcription of the vector's transgene as well as the other genes in the vector such as the antibiotic resistance gene. Some cloning vectors need not have a promoter for the cloned insert but it is an essential component of expression vectors so that the cloned product may be expressed.
- Cloning site: This may be a multiple cloning site or other features that allow for the insertion of foreign DNA into the vector through ligation.
- Genetic markers: Genetic markers for viral vectors allow for confirmation that the vector has integrated with the host genomic DNA.
- Antibiotic resistance: Vectors with antibiotic-resistance open reading frames allow for survival of cells that have taken up the vector in growth media containing antibiotics through antibiotic selection.
- Epitope: Some vectors may contain a sequence for a specific epitope that can be incorporated into the expressed protein. It allows for antibody identification of cells expressing the target protein.
- Reporter genes: Some vectors may contain a reporter gene that allow for identification of plasmid that contains inserted DNA sequence. An example is lacZ-α which codes for the N-terminus fragment of β-galactosidase, an enzyme that digests galactose. A multiple cloning site is located within lacZ-α, and an insert successfully ligated into the vector will disrupt the gene sequence, resulting in an inactive β-galactosidase. Cells containing vector with an insert may be identified using blue/white selection by growing cells in media containing an analogue of galactose (X-gal). Cells expressing β-galactosidase (therefore doesn't contain an insert) appear as blue colonies. White colonies would be selected as those that may contain an insert. Other commonly used reporters include green fluorescent protein and luciferase.
- Targeting sequence: Expression vectors may include encoding for a targeting sequence in the finished protein that directs the expressed protein to a specific organelle in the cell or specific location such as the periplasmic space of bacteria.
- Protein purification tags: Some expression vectors include proteins or peptide sequences that allows for easier purification of the expressed protein. Examples include polyhistidine-tag, glutathione-S-transferase, and maltose binding protein. Some of these tags may also allow for increased solubility of the target protein. The target protein is fused to the protein tag, but a protease cleavage site positioned in the polypeptide linker region between the protein and the tag allows the tag to be removed later.
See also
References
- Lodish H, Berk A, Zipursky SL, Matsudaira P, Baltimore D, Darnell J (2000). "DNA Cloning with Plasmid Vectors". Molecular Cell Biology (4th ed.). New York: W. H. Freeman.
- Acquaah G (16 August 2012). Principles of Plant Genetics and Breeding. John Wiley & Sons Inc. ISBN 978-1-118-31369-5.
- Johnston C, Martin B, Fichant G, Polard P, Claverys JP (March 2014). "Bacterial transformation: distribution, shared mechanisms and divergent control". Nature Reviews. Microbiology. 12 (3): 181–96. doi:10.1038/nrmicro3199. PMID 24509783. S2CID 23559881.
- "MeSH Browser". meshb.nlm.nih.gov. Retrieved 2018-04-16.
- Hartl DL, Jones EW (1998). Genetics: principles and analysis (4th ed.). Sudbury, Mass.: Jones and Bartlett Publishers. ISBN 978-0-7637-0489-6. OCLC 45730915.
- del Solar, Gloria; Giraldo, Rafael; Ruiz-Echevarría, María Jesús; Espinosa, Manuel; Díaz-Orejas, Ramón (June 1998). "Replication and Control of Circular Bacterial Plasmids". Microbiology and Molecular Biology Reviews. 62 (2): 434–464. doi:10.1128/MMBR.62.2.434-464.1998. ISSN 1092-2172. PMC 98921. PMID 9618448.
- Brown TA (2010). "Chapter 2 - Vectors for Gene Cloning: Plasmids and Bacteriophages". Gene Cloning and DNA Analysis: An Introduction (6th ed.). Wiley-Blackwell. ISBN 978-1-4051-8173-0.
- Julin, Douglas (2014). "Artificial Chromosomes". Molecular Life Sciences. Springer, New York, NY. pp. 1–3. doi:10.1007/978-1-4614-6436-5_91-3. ISBN 978-1-4614-6436-5.
- Murray, Andrew; Szostak, Jack (November 1987). "Artificial Chromosomes". Scientific American. 257 (5): 62–68. Bibcode:1987SciAm.257e..62M. doi:10.1038/scientificamerican1187-62. PMID 3317814.
- Solomon EP, Berg LR, Martin DW (2005). Biology (8th ed.). Belmont, CA: Brooks/Cole Thomson Learning. ISBN 978-0-495-31714-2. OCLC 123008833.
- Damdindorj L, Karnan S, Ota A, Hossain E, Konishi Y, Hosokawa Y, Konishi H (2014-08-29). "A comparative analysis of constitutive promoters located in adeno-associated viral vectors". PLOS ONE. 9 (8): e106472. Bibcode:2014PLoSO...9j6472D. doi:10.1371/journal.pone.0106472. PMC 4149579. PMID 25170953.
- Lewin A, Mayer M, Chusainow J, Jacob D, Appel B (June 2005). "Viral promoters can initiate expression of toxin genes introduced into Escherichia coli". BMC Biotechnology. 5: 19. doi:10.1186/1472-6750-5-19. PMC 1181807. PMID 15967027.
Further reading
- Freshney IR (2005-07-29). Culture of Animal Cells: A manual of basic technique. Hoboken, New Jersey: John Wiley & Sons, Inc. ISBN 978-0-471-45329-1.