Vitamin D-binding protein
Vitamin D-binding protein (DBP), also/originally known as gc-globulin (group-specific component), is a protein that in humans is encoded by the GC gene.[5][6]
Structure
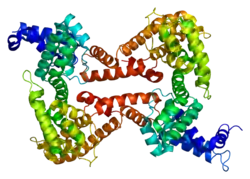
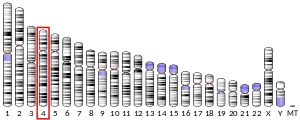
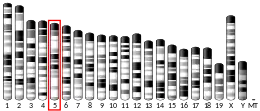
Human GC is a glycosylated alpha-globulin, ~58 kDa in size. Its 458 amino acids are coded for by 1690 nucleotides on chromosome 4 (4q11–q13). The primary structure contains 28 cysteine residues forming multiple disulfide bonds. GC contains 3 domains. Domain 1 is composed of 10 alpha helices, domain 2 of 9, and domain 3 of 4.[7]
Function
Vitamin D-binding protein belongs to the albumin gene family, together with human serum albumin and alpha-fetoprotein. It is a multifunctional protein found in plasma, ascitic fluid, cerebrospinal fluid and on the surface of many cell types.
It is able to bind the various forms of vitamin D including ergocalciferol (vitamin D2) and cholecalciferol (vitamin D3), the 25-hydroxylated forms (calcifediol), and the active hormonal product, 1,25-dihydroxyvitamin D (calcitriol). The major proportion of vitamin D in blood is bound to this protein. It transports vitamin D metabolites between skin, liver and kidney, and then on to the various target tissues.[6][8]
As Gc protein-derived macrophage activating factor it is a Macrophage Activating Factor (MAF) that has been tested for use as a cancer treatment that would activate macrophages against cancer cells.[9]
Interactive pathway map
Click on genes, proteins and metabolites below to link to respective articles. [§ 1]
- The interactive pathway map can be edited at WikiPathways: "VitaminDSynthesis_WP1531".
Production
It is synthesized by hepatic parenchymal cells and secreted into the blood circulation.[8]
Genetic variation
Many genetic variants of the GC gene are known. They produce 6 main haplotypes and 3 main protein variants (Gc1S, Gc1F and Gc2).[10] The genetic variations are associated with differences in circulating 25-hydroxyvitamin D levels.[11] They have been proposed to account for some of the differences in vitamin D status in different ethnic groups,[12] and have been found to correlate with the response to vitamin D supplementation.[10]
References
- GRCh38: Ensembl release 89: ENSG00000145321 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000035540 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Mikkelsen M, Jacobsen P, Henningsen K (Jul 1977). "Possible localization of Gc-System on chromosome 4. Loss of long arm 4 material associated with father-child incompatibility within the Gc-System". Human Heredity. 27 (2): 105–7. doi:10.1159/000152857. PMID 558959.
- "Entrez Gene: GC group-specific component (vitamin D binding protein)".
- Verboven C, Rabijns A, De Maeyer M, Van Baelen H, Bouillon R, De Ranter C (February 2002). "A structural basis for the unique binding features of the human vitamin D-binding protein". Nature Structural Biology. 9 (2): 131–6. doi:10.1038/nsb754. PMID 11799400. S2CID 38990672.
- Norman AW (August 2008). "From vitamin D to hormone D: fundamentals of the vitamin D endocrine system essential for good health". The American Journal of Clinical Nutrition. 88 (2): 491S–499S. doi:10.1093/ajcn/88.2.491S. PMID 18689389.
- Yamamoto N, Suyama H, Yamamoto N (July 2008). "Immunotherapy for Prostate Cancer with Gc Protein-Derived Macrophage-Activating Factor, GcMAF" ([PDF]). Translational Oncology. 1 (2): 65–72. doi:10.1593/tlo.08106. PMC 2510818. PMID 18633461.
- Malik S, Fu L, Juras DJ, Karmali M, Wong BY, Gozdzik A, Cole DE (January–February 2013). "Common variants of the vitamin D binding protein gene and adverse health outcomes". Critical Reviews in Clinical Laboratory Sciences. 50 (1): 1–22. doi:10.3109/10408363.2012.750262. PMC 3613945. PMID 23427793.
- McGrath JJ, Saha S, Burne TH, Eyles DW (July 2010). "A systematic review of the association between common single nucleotide polymorphisms and 25-hydroxyvitamin D concentrations". The Journal of Steroid Biochemistry and Molecular Biology. 121 (1–2): 471–7. doi:10.1016/j.jsbmb.2010.03.073. PMID 20363324. S2CID 20057294.
- Powe CE, Evans MK, Wenger J, Zonderman AB, Berg AH, Nalls M, Tamez H, Zhang D, Bhan I, Karumanchi SA, Powe NR, Thadhani R (November 2013). "Vitamin D-binding protein and vitamin D status of black Americans and white Americans". The New England Journal of Medicine. 369 (21): 1991–2000. doi:10.1056/NEJMoa1306357. PMC 4030388. PMID 24256378.
Further reading
- Svasti J, Kurosky A, Bennett A, Bowman BH (April 1979). "Molecular basis for the three major forms of human serum vitamin D binding protein (group-specific component)". Biochemistry. 18 (8): 1611–7. doi:10.1021/bi00575a036. PMID 218624.
- Braun A, Bichlmaier R, Cleve H (June 1992). "Molecular analysis of the gene for the human vitamin-D-binding protein (group-specific component): allelic differences of the common genetic GC types". Human Genetics. 89 (4): 401–6. doi:10.1007/BF00194311. PMID 1352271. S2CID 1913655.
- Esteban C, Geuskens M, Ena JM, Mishal Z, Macho A, Torres JM, Uriel J (May 1992). "Receptor-mediated uptake and processing of vitamin D-binding protein in human B-lymphoid cells". The Journal of Biological Chemistry. 267 (14): 10177–83. PMID 1374401.
- Szpirer C, Riviere M, Cortese R, Nakamura T, Islam MQ, Levan G, Szpirer J (June 1992). "Chromosomal localization in man and rat of the genes encoding the liver-enriched transcription factors C/EBP, DBP, and HNF1/LFB-1 (CEBP, DBP, and transcription factor 1, TCF1, respectively) and of the hepatocyte growth factor/scatter factor gene (HGF)". Genomics. 13 (2): 293–300. doi:10.1016/0888-7543(92)90245-N. PMID 1535333.
- Dawson SJ, White LA (May 1992). "Treatment of Haemophilus aphrophilus endocarditis with ciprofloxacin". The Journal of Infection. 24 (3): 317–20. doi:10.1016/S0163-4453(05)80037-4. PMID 1602151.
- Yang F, Bergeron JM, Linehan LA, Lalley PA, Sakaguchi AY, Bowman BH (August 1990). "Mapping and conservation of the group-specific component gene in mouse". Genomics. 7 (4): 509–16. doi:10.1016/0888-7543(90)90193-X. PMID 1696927.
- Yang F, Luna VJ, McAnelly RD, Naberhaus KH, Cupples RL, Bowman BH (November 1985). "Evolutionary and structural relationships among the group-specific component, albumin and alpha-fetoprotein". Nucleic Acids Research. 13 (22): 8007–17. doi:10.1093/nar/13.22.8007. PMC 322106. PMID 2415926.
- Yang F, Brune JL, Naylor SL, Cupples RL, Naberhaus KH, Bowman BH (December 1985). "Human group-specific component (Gc) is a member of the albumin family". Proceedings of the National Academy of Sciences of the United States of America. 82 (23): 7994–8. doi:10.1073/pnas.82.23.7994. PMC 391428. PMID 2415977.
- Cooke NE, David EV (December 1985). "Serum vitamin D-binding protein is a third member of the albumin and alpha fetoprotein gene family". The Journal of Clinical Investigation. 76 (6): 2420–4. doi:10.1172/JCI112256. PMC 424397. PMID 2416779.
- Schoentgen F, Metz-Boutigue MH, Jollès J, Constans J, Jollès P (June 1986). "Complete amino acid sequence of human vitamin D-binding protein (group-specific component): evidence of a three-fold internal homology as in serum albumin and alpha-fetoprotein". Biochimica et Biophysica Acta (BBA) - Protein Structure and Molecular Enzymology. 871 (2): 189–98. doi:10.1016/0167-4838(86)90173-1. PMID 2423133.
- McNearney TA, Odell C, Holers VM, Spear PG, Atkinson JP (November 1987). "Herpes simplex virus glycoproteins gC-1 and gC-2 bind to the third component of complement and provide protection against complement-mediated neutralization of viral infectivity". The Journal of Experimental Medicine. 166 (5): 1525–35. doi:10.1084/jem.166.5.1525. PMC 2189652. PMID 2824652.
- Yang F, Naberhaus KH, Adrian GS, Gardella JM, Brissenden JE, Bowman BH (1987). "The vitamin D-binding protein gene contains conserved nucleotide sequences that respond to heavy metal, adipocyte and mitotic signals". Gene. 54 (2–3): 285–90. doi:10.1016/0378-1119(87)90499-9. PMID 2958390.
- Cooke NE, Willard HF, David EV, George DL (July 1986). "Direct regional assignment of the gene for vitamin D binding protein (Gc-globulin) to human chromosome 4q11-q13 and identification of an associated DNA polymorphism". Human Genetics. 73 (3): 225–9. doi:10.1007/BF00401232. PMID 3015768. S2CID 38816588.
- Nestler JE, McLeod JF, Kowalski MA, Strauss JF, Haddad JG (May 1987). "Detection of vitamin D binding protein on the surface of cytotrophoblasts isolated from human placentae". Endocrinology. 120 (5): 1996–2002. doi:10.1210/endo-120-5-1996. PMID 3552627.
- Pierce EA, Dame MC, Bouillon R, Van Baelen H, DeLuca HF (December 1985). "Monoclonal antibodies to human vitamin D-binding protein". Proceedings of the National Academy of Sciences of the United States of America. 82 (24): 8429–33. doi:10.1073/pnas.82.24.8429. PMC 390929. PMID 3936035.
- Wooten MW, Nel AE, Goldschmidt-Clermont PJ, Galbraith RM, Wrenn RW (October 1985). "Identification of a major endogenous substrate for phospholipid/Ca2+-dependent kinase in pancreatic acini as Gc (vitamin D-binding protein)". FEBS Letters. 191 (1): 97–101. doi:10.1016/0014-5793(85)81001-2. PMID 4054306. S2CID 29613843.
- Constans J, Oksman F, Viau M (August 1981). "Binding of the apo and holo forms of the serum vitamin D-binding protein to human lymphocyte cytoplasm and membrane by indirect immunofluorescence". Immunology Letters. 3 (3): 159–62. doi:10.1016/0165-2478(81)90120-6. PMID 7026425.
- Braun A, Kofler A, Morawietz S, Cleve H (December 1993). "Sequence and organization of the human vitamin D-binding protein gene". Biochimica et Biophysica Acta (BBA) - Gene Structure and Expression. 1216 (3): 385–94. doi:10.1016/0167-4781(93)90005-x. PMID 7505619.
- Swamy N, Roy A, Chang R, Brisson M, Ray R (April 1995). "Affinity purification of human plasma vitamin D-binding protein". Protein Expression and Purification. 6 (2): 185–8. doi:10.1006/prep.1995.1023. PMID 7606167.
External links
- Overview of all the structural information available in the PDB for UniProt: P02774 (Vitamin D-binding protein) at the PDBe-KB.