Antigen
In immunology, an antigen (Ag) is a molecule or molecular structure, such as may be present on the outside of a pathogen, that can be bound by an antigen-specific antibody or B-cell antigen receptor.[1] The presence of antigens in the body normally triggers an immune response.[2] The Ag abbreviation stands for an antibody generator.[3]
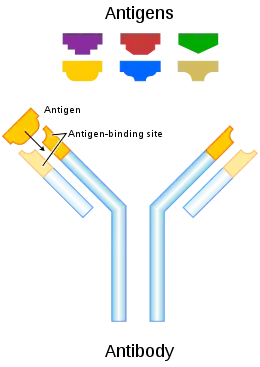
Antigens are "targeted" by antibodies.[1] Each antibody is specifically produced by the immune system to match an antigen after cells in the immune system come into contact with it; this allows a precise identification or matching of the antigen and the initiation of an adaptive response.[1][2] The antibody is said to "match" the antigen in the sense that it can bind to it due to an adaptation in a antigen-binding fragment of the antibody.[1] In most cases, an adapted antibody can only react to and bind one specific antigen; in some instances, however, antibodies may cross-react and bind more than one antigen.
Antigens are proteins, peptides (amino acid chains) and polysaccharides (chains of monosaccharides/simple sugars) but lipids and nucleic acids become antigens only when combined with proteins and polysaccharides.[4]
The antigen may originate from within the body ("self-antigen") or from the external environment ("non-self").[2] The immune system identifies and attacks "non-self" external antigens and usually does not react to self-antigens due to negative selection of T cells in the thymus.[5]
Vaccines are examples of antigens in an immunogenic form, which are intentionally administered to a recipient to induce the memory function of the adaptive immune system towards antigens of the pathogen invading that recipient. Vaccines for the seasonal flu virus is a common example.[6]
Etymology
Paul Ehrlich coined the term antibody (in German Antikörper) in his side-chain theory at the end of the 19th century.[7] In 1899, Ladislas Deutsch (László Detre) (1874–1939) named the hypothetical substances halfway between bacterial constituents and antibodies "substances immunogenes ou antigenes" (antigenic or immunogenic substances). He originally believed those substances to be precursors of antibodies, just as zymogen is a precursor of an enzyme. But, by 1903, he understood that an antigen induces the production of immune bodies (antibodies) and wrote that the word antigen is a contraction of antisomatogen (Immunkörperbildner). The Oxford English Dictionary indicates that the logical construction should be "anti(body)-gen".[8]
Terminology
- Epitope – the distinct surface features of an antigen, its antigenic determinant.
Antigenic molecules, normally "large" biological polymers, usually present surface features that can act as points of interaction for specific antibodies. Any such feature constitutes an epitope. Most antigens have the potential to be bound by multiple antibodies, each of which is specific to one of the antigen's epitopes. Using the "lock and key" metaphor, the antigen can be seen as a string of keys (epitopes) each of which matches a different lock (antibody). Different antibody idiotypes, each have distinctly formed complementarity-determining regions.
- Allergen – A substance capable of causing an allergic reaction .The (detrimental) reaction may result after exposure via ingestion, inhalation, injection, or contact with skin.
- Superantigen – A class of antigens that cause non-specific activation of T-cells, resulting in polyclonal T-cell activation and massive cytokine release.
- Tolerogen – A substance that invokes a specific immune non-responsiveness due to its molecular form. If its molecular form is changed, a tolerogen can become an immunogen.
- Immunoglobulin-binding protein – Proteins such as protein A, protein G, and protein L that are capable of binding to antibodies at positions outside of the antigen-binding site. While antigens are the "target" of antibodies, immunoglobulin-binding proteins "attack" antibodies.
- T-dependent antigen – Antigens that require the assistance of T cells to induce the formation of specific antibodies.
- T-independent antigen – Antigens that stimulate B cells directly.
- Immunodominant antigens – Antigens that dominate (over all others from a pathogen) in their ability to produce an immune response. T cell responses typically are directed against a relatively few immunodominant epitopes, although in some cases (e.g., infection with the malaria pathogen Plasmodium spp.) it is dispersed over a relatively large number of parasite antigens.[9]
Antigen-presenting cells present antigens in the form of peptides on histocompatibility molecules. The T cells selectively recognize the antigens; depending on the antigen and the type of the histocompatibility molecule, different types of T cells will be activated. For T-cell receptor (TCR) recognition, the peptide must be processed into small fragments inside the cell and presented by a major histocompatibility complex (MHC).[10] The antigen cannot elicit the immune response without the help of an immunologic adjuvant.[4] Similarly, the adjuvant component of vaccines plays an essential role in the activation of the innate immune system.[11][12]
An immunogen is an antigen substance (or adduct) that is able to trigger a humoral (innate) or cell-mediated immune response.[13] It first initiates an innate immune response, which then causes the activation of the adaptive immune response. An antigen binds the highly variable immunoreceptor products (B-cell receptor or T-cell receptor) once these have been generated. Immunogens are those antigens, termed immunogenic, capable of inducing an immune response.[14]
At the molecular level, an antigen can be characterized by its ability to bind to an antibody's variable Fab region. Different antibodies have the potential to discriminate among specific epitopes present on the antigen surface. A hapten is a small molecule that changes the structure of an antigenic epitope. In order to induce an immune response, it needs to be attached to a large carrier molecule such as a protein (a complex of peptides). Antigens are usually carried by proteins and polysaccharides, and less frequently, lipids. This includes parts (coats, capsules, cell walls, flagella, fimbriae, and toxins) of bacteria, viruses, and other microorganisms. Lipids and nucleic acids are antigenic only when combined with proteins and polysaccharides. Non-microbial non-self antigens can include pollen, egg white, and proteins from transplanted tissues and organs or on the surface of transfused blood cells.
Sources
Antigens can be classified according to their source.
Exogenous antigens
Exogenous antigens are antigens that have entered the body from the outside, for example, by inhalation, ingestion or injection. The immune system's response to exogenous antigens is often subclinical. By endocytosis or phagocytosis, exogenous antigens are taken into the antigen-presenting cells (APCs) and processed into fragments. APCs then present the fragments to T helper cells (CD4+) by the use of class II histocompatibility molecules on their surface. Some T cells are specific for the peptide:MHC complex. They become activated and start to secrete cytokines, substances that activate cytotoxic T lymphocytes (CTL), antibody-secreting B cells, macrophages and other particles.
Some antigens start out as exogenous and later become endogenous (for example, intracellular viruses). Intracellular antigens can be returned to circulation upon the destruction of the infected cell.
Endogenous antigens
Endogenous antigens are generated within normal cells as a result of normal cell metabolism, or because of viral or intracellular bacterial infection. The fragments are then presented on the cell surface in the complex with MHC class I molecules. If activated cytotoxic CD8+ T cells recognize them, the T cells secrete various toxins that cause the lysis or apoptosis of the infected cell. In order to keep the cytotoxic cells from killing cells just for presenting self-proteins, the cytotoxic cells (self-reactive T cells) are deleted as a result of tolerance (negative selection). Endogenous antigens include xenogenic (heterologous), autologous and idiotypic or allogenic (homologous) antigens. Sometimes antigens are part of the host itself in an autoimmune disease.[2]
Autoantigens
An autoantigen is usually a normal protein or protein complex (and sometimes DNA or RNA) that is recognized by the immune system of patients suffering from a specific autoimmune disease. Under normal conditions, these antigens should not be the target of the immune system, but in autoimmune diseases, their associated T cells are not deleted and instead attack.
Neoantigens
Neoantigens are those that are entirely absent from the normal human genome. As compared with nonmutated self-antigens, neoantigens are of relevance to tumor control, as the quality of the T cell pool that is available for these antigens is not affected by central T cell tolerance. Technology to systematically analyze T cell reactivity against neoantigens became available only recently.[15] Neoantigens can be directly detected and quantified through a method called MANA-SRM developed by a molecular diagnostics company, Complete Omics Inc., through collaborating with a team in Johns Hopkins University School of Medicine. [16]
Viral antigens
For virus-associated tumors, such as cervical cancer and a subset of head and neck cancers, epitopes derived from viral open reading frames contribute to the pool of neoantigens.[15]
Tumor antigens
Tumor antigens are those antigens that are presented by MHC class I or MHC class II molecules on the surface of tumor cells. Antigens found only on such cells are called tumor-specific antigens (TSAs) and generally result from a tumor-specific mutation. More common are antigens that are presented by tumor cells and normal cells, called tumor-associated antigens (TAAs). Cytotoxic T lymphocytes that recognize these antigens may be able to destroy tumor cells.[15]
Tumor antigens can appear on the surface of the tumor in the form of, for example, a mutated receptor, in which case they are recognized by B cells.[15]
For human tumors without a viral etiology, novel peptides (neo-epitopes) are created by tumor-specific DNA alterations.[15]
Process
A large fraction of human tumor mutations is effectively patient-specific. Therefore, neoantigens may also be based on individual tumor genomes. Deep-sequencing technologies can identify mutations within the protein-coding part of the genome (the exome) and predict potential neoantigens. In mice models, for all novel protein sequences, potential MHC-binding peptides were predicted. The resulting set of potential neoantigens was used to assess T cell reactivity. Exome–based analyses were exploited in a clinical setting, to assess reactivity in patients treated by either tumor-infiltrating lymphocyte (TIL) cell therapy or checkpoint blockade. Neoantigen identification was successful for multiple experimental model systems and human malignancies.[15]
The false-negative rate of cancer exome sequencing is low—i.e.: the majority of neoantigens occur within exonic sequence with sufficient coverage. However, the vast majority of mutations within expressed genes do not produce neoantigens that are recognized by autologous T cells.[15]
As of 2015 mass spectrometry resolution is insufficient to exclude many false positives from the pool of peptides that may be presented by MHC molecules. Instead, algorithms are used to identify the most likely candidates. These algorithms consider factors such as the likelihood of proteasomal processing, transport into the endoplasmic reticulum, affinity for the relevant MHC class I alleles and gene expression or protein translation levels.[15]
The majority of human neoantigens identified in unbiased screens display a high predicted MHC binding affinity. Minor histocompatibility antigens, a conceptually similar antigen class are also correctly identified by MHC binding algorithms. Another potential filter examines whether the mutation is expected to improve MHC binding. The nature of the central TCR-exposed residues of MHC-bound peptides is associated with peptide immunogenicity.[15]
Antigenic specificity
Antigenic specificity is the ability of the host cells to recognize an antigen specifically as a unique molecular entity and distinguish it from another with exquisite precision. Antigen specificity is due primarily to the side-chain conformations of the antigen. It is measurable and need not be linear or of a rate-limited step or equation.[2][6] Both T cells and B cells are cellular components of adaptive immunity.[2][17]
See also
References
- "Antibody". National Human Genome Research Institute, US National Institutes of Health. 2020. Retrieved 13 October 2020.
- "Immune system and disorders". MedlinePlus, US National Institute of Medicine. 28 September 2020. Retrieved 13 October 2020.
- Male, David K. (2006). Immunology. Elsevier Health Sciences. p. 10. ISBN 978-0323033992.
- Gavin, AL; Hoebe, K; Duong, B; Ota, T; Martin, C; Beutler, B; Nemazee, D (22 December 2006). "Adjuvant-enhanced antibody responses in the absence of toll-like receptor signaling". Science. 314 (5807): 1936–38. Bibcode:2006Sci...314.1936G. doi:10.1126/science.1135299. PMC 1868398. PMID 17185603.
- Gallucci, S; Lolkema, M; Matzinger, P (November 1999). "Natural adjuvants: endogenous activators of dendritic cells". Nature Medicine. 5 (11): 1249–55. doi:10.1038/15200. PMID 10545990. S2CID 29090284.
- "Antigenic characterization". US Centers for Disease Control and Prevention. 15 October 2019. Retrieved 13 October 2020.
- Strebhardt, Klaus; Ullrich, Axel (Jun 2008). "Paul Ehrlich's magic bullet concept: 100 years of progress". Nature Reviews Cancer. 8 (6): 473–80. doi:10.1038/nrc2394. ISSN 1474-1768. PMID 18469827. S2CID 30063909.
- Lindenmann, Jean (1984). "Origin of the Terms 'Antibody' and 'Antigen'". Scand. J. Immunol. 19 (4): 281–85. doi:10.1111/j.1365-3083.1984.tb00931.x. PMID 6374880. Retrieved 2008-10-31.
- Doolan DL, Southwood S, Freilich DA, Sidney J, Graber NL, Shatney L, Bebris L, Florens L, Dobano C, Witney AA, Appella E, Hoffman SL, Yates JR, Carucci DJ, Sette A (August 2003). "Identification of Plasmodium falciparum antigens by antigenic analysis of genomic and proteomic data". Proceedings of the National Academy of Sciences of the United States of America. 100 (17): 9952–57. Bibcode:2003PNAS..100.9952D. doi:10.1073/pnas.1633254100. PMC 187898. PMID 12886016.
- Parham, Peter. (2009). The Immune System, 3rd Edition, p. G:2, Garland Science, Taylor and Francis Group, LLC.
- Janeway CA, Jr (1 November 2013). "Pillars article: approaching the asymptote? Evolution and revolution in immunology. Cold spring harb symp quant biol. 1989. 54: 1–13". Journal of Immunology. 191 (9): 4475–87. PMID 24141854.
- Gayed, PM (June 2011). "Toward a modern synthesis of immunity: Charles A. Janeway Jr. and the immunologist's dirty little secret". The Yale Journal of Biology and Medicine. 84 (2): 131–38. ISSN 1551-4056. PMC 3117407. PMID 21698045.
- Parham, Peter. (2009). The Immune System, 3rd Edition, p. G:11, Garland Science, Taylor and Francis Group, LLC.
- Kuby Immunology (6th ed.). Macmillan. 2006. p. 77. ISBN 978-1-4292-0211-4.
- Schumacher, Ton N.; Schreiber, Robert D. (April 3, 2015). "Neoantigens in cancer immunotherapy". Science. 348 (6230): 69–74. Bibcode:2015Sci...348...69S. doi:10.1126/science.aaa4971. PMID 25838375.
- Wang, Qing.; Douglass, Jacqueline (September 16, 2019). "Direct Detection and Quantification of Neoantigens". Cancer Immunol Res. 7 (11): 1748–54. doi:10.1158/2326-6066.CIR-19-0107. PMC 6825591. PMID 31527070.
- K. Abbas, Abul; Lichtman, Andrew; Pillai, Shiv (2018). Cellular and molecular immunology (Ninth ed.). Philadelphia: Elsevier. p. 97. ISBN 978-0-323-52324-0.