D-Amino acid
D-Amino acids are amino acids where the stereogenic carbon alpha to the amino group has the D-configuration. For most naturally-occurring amino acids, this carbon has the L-configuration. D-Amino acids are occasionally found in nature as residues in proteins. They are formed from ribosomally-derived D-amino acid residues.[1]
Structure and general properties
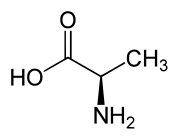 D-alanine.
D-alanine.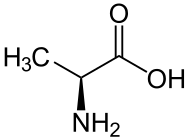 L-alanine.
L-alanine.
L- and D-amino acids are usually enantiomers. The exceptions are two amino acids with two stereogenic centers, threonine and isoleucine. Aside from those two special cases, L- and D-amino acids have identical properties (color, solubility, melting point) under many conditions. In the biological context however, which is chiral, these enantiomers can behave very differently. Thus, D-amino acids have low nutritional value, in part because they are not digested well.[2]
Occurrence and use
D-amino acid residues occur in cone snails.[3] They are also abundant components of the peptidoglycan cell walls of bacteria,[4] and D-serine may act as a neurotransmitter in the brain.[5] D-amino acids are used in racemic crystallography to create centrosymmetric crystals, which, depending on the protein, may allow for easier and more robust protein structure determination.[6]
Gramicidin is a polypeptide made up from mixture of D- and L-amino acids.[7] Other compounds containing D-amino acids are tyrocidine and valinomycin. These compounds disrupt bacterial cell walls, particularly in Gram-positive bacteria. As of 2011, only 837 D-amino acids were found in the Swiss-Prot database out of a total of 187 million amino acids analysed.[8]
Biosynthesis
Two enzymes convert L-amino acids to D-amino acids. D-Amino-acid racemase, a PLP-dependent enzyme, racemizes amino acids via the formation of the alpha-iminoacids, where the stereogenic center is lost. L-amino-acid oxidases convert L-amino acids to the alpha-ketoacids, which are susceptible to reductive amination. Some amino acids are prone to racemization, one example being lysine, which racemizes via formation of pipecolic acid.
In peptides, L-amino acid residues slowly racemize, resulting in the formation of some D-amino acid residues. Racemization occurs via deprotonation of the methyne that is alpha to the amido group. Rates increase with pH.
Many D-amino acids found in higher organisms are derived from microbial sources. The D-alanine in peptidoglycans that comprise bacterial cell walls helps its host resist attack by proteolytic enzymes. Several antibiotics, e.g. bacitracin, contain D-amino acid residues.[2]
References
- Genchi, Giuseppe (2017). "An overview on d-amino acids". Amino Acids. 49 (9): 1521–1533. doi:10.1007/s00726-017-2459-5. PMID 28681245. S2CID 3998765.
- Mendel Friedman (1999). "Chemistry, Nutrition, and Microbiology of D-Amino Acids". J. Agric. Food Chem. 47 (9): 3457–79. doi:10.1021/jf990080u. PMID 10552672.
- Pisarewicz K, Mora D, Pflueger FC, Fields GB, Marí F (May 2005). "Polypeptide chains containing D-gamma-hydroxyvaline". Journal of the American Chemical Society. 127 (17): 6207–6215. doi:10.1021/ja050088m. PMID 15853325.
- van Heijenoort J (March 2001). "Formation of the glycan chains in the synthesis of bacterial peptidoglycan". Glycobiology. 11 (3): 25R–36R. doi:10.1093/glycob/11.3.25R. PMID 11320055. S2CID 46066256.
- Wolosker H, Dumin E, Balan L, Foltyn VN (July 2008). "D-Amino acids in the brain: D-serine in neurotransmission and neurodegeneration". The FEBS Journal. 275 (14): 3514–3526. doi:10.1111/j.1742-4658.2008.06515.x. PMID 18564180. S2CID 25735605.
- Matthews BW (June 2009). "Racemic crystallography—easy crystals and easy structures: What's not to like?". Protein Science. 18 (6): 1135–1138. doi:10.1002/pro.125. PMC 2774423. PMID 19472321.
- Ketchem RR, Hu W, Cross TA (September 1993). "High-resolution conformation of gramicidin A in a lipid bilayer by solid-state NMR". Science. 261 (5127): 1457–1460. Bibcode:1993Sci...261.1457K. doi:10.1126/science.7690158. PMID 7690158.
- Khoury GA, Baliban RC, Floudas CA (September 2011). "Proteome-wide post-translational modification statistics: frequency analysis and curation of the swiss-prot database". Scientific Reports. 1 (90): 90. Bibcode:2011NatSR...1E..90K. doi:10.1038/srep00090. PMC 3201773. PMID 22034591.