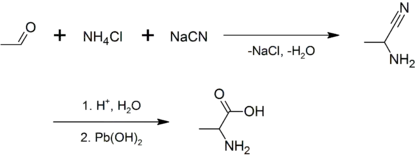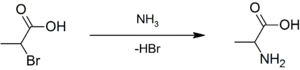Alanine
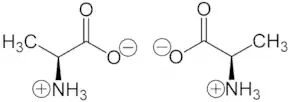
Alanine (symbol Ala or A)[3] is an α-amino acid that is used in the biosynthesis of proteins. It contains an amine group and a carboxylic acid group, both attached to the central carbon atom which also carries a methyl group side chain. Consequently, its IUPAC systematic name is 2-aminopropanoic acid, and it is classified as a nonpolar, aliphatic α-amino acid. Under biological conditions, it exists in its zwitterionic form with its amine group protonated (as −NH3+) and its carboxyl group deprotonated (as −CO2−). It is non-essential to humans as it can be synthesised metabolically and does not need to be present in the diet. It is encoded by all codons starting with GC (GCU, GCC, GCA, and GCG).
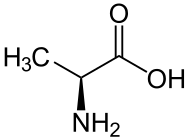 Skeletal formula of L-alanine | |||
| |||
| Names | |||
|---|---|---|---|
| IUPAC name
Alanine | |||
| Other names
2-Aminopropanoic acid | |||
| Identifiers | |||
3D model (JSmol) |
| ||
| 3DMet | |||
| 1720248 | |||
| ChEBI | |||
| ChEMBL | |||
| ChemSpider | |||
| DrugBank | |||
| ECHA InfoCard | 100.000.249 | ||
| EC Number |
| ||
| 49628 | |||
| KEGG | |||
PubChem CID |
|||
| UNII |
| ||
CompTox Dashboard (EPA) |
|||
| |||
| |||
| Properties | |||
| C3H7NO2 | |||
| Molar mass | 89.094 g·mol−1 | ||
| Appearance | white powder | ||
| Density | 1.424 g/cm3 | ||
| Melting point | 258 °C (496 °F; 531 K) (sublimes) | ||
| 167.2 g/L (25 °C) | |||
| log P | -0.68[1] | ||
| Acidity (pKa) |
| ||
| -50.5·10−6 cm3/mol | |||
| Supplementary data page | |||
| Refractive index (n), Dielectric constant (εr), etc. | |||
Thermodynamic data |
Phase behaviour solid–liquid–gas | ||
| UV, IR, NMR, MS | |||
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa). | |||
| Infobox references | |||
The L-isomer of alanine (left-handed) is the one that is incorporated into proteins. L-Alanine is second only to leucine in rate of occurrence, accounting for 7.8% of the primary structure in a sample of 1,150 proteins.[4] The right-handed form, D-alanine, occurs in polypeptides in some bacterial cell walls[5]:131 and in some peptide antibiotics, and occurs in the tissues of many crustaceans and molluscs as an osmolyte.[6] Alanine is also very abundant (over-represented) in low complexity regions of proteins.[7]
History and etymology
Alanine was first synthesized in 1850 when Adolph Strecker combined acetaldehyde and ammonia with hydrogen cyanide.[8][9][10] The amino acid was named Alanin in German, in reference to aldehyde, with the infix -an- for ease of pronunciation,[11] the German ending -in used in chemical compounds being analogous to English -ine.
Structure
Alanine is an aliphatic amino acid, because the side-chain connected to the α-carbon atom is a methyl group (-CH3); alanine is the simplest α-amino acid after glycine. The methyl side-chain of alanine is non-reactive and is therefore hardly ever directly involved in protein function.[12] Alanine is a nonessential amino acid, meaning it can be manufactured by the human body, and does not need to be obtained through the diet. Alanine is found in a wide variety of foods, but is particularly concentrated in meats.
Sources
Biosynthesis
Alanine can be synthesized from pyruvate and branched chain amino acids such as valine, leucine, and isoleucine.
Alanine is produced by reductive amination of pyruvate, a two-step process. In the first step, α-ketoglutarate, ammonia and NADH are converted by glutamate dehydrogenase to glutamate, NAD+ and water. In the second step, the amino group of the newly-formed glutamate is transferred to pyruvate by an aminotransferase enzyme, regenerating the α-ketoglutarate, and converting the pyruvate to alanine. The net result is that pyruvate and ammonia are converted to alanine, consuming one reducing equivalent.[5]:721 Because transamination reactions are readily reversible and pyruvate is present in all cells, alanine can be easily formed and thus has close links to metabolic pathways such as glycolysis, gluconeogenesis, and the citric acid cycle.
Chemical synthesis
L-Alanine is produced industrially by decarboxylation of L-aspartate by the action of aspartate 4-decarboxylase. Fermentation routes to L-alanine are complicated by alanine racemase.[13]
Racemic alanine can be prepared by the condensation of acetaldehyde with ammonium chloride in the presence of sodium cyanide by the Strecker reaction,[14] or by the ammonolysis of 2-bromopropanoic acid.[15]
Degradation
Alanine is broken down by oxidative deamination, the inverse reaction of the reductive amination reaction described above, catalyzed by the same enzymes. The direction of the process is largely controlled by the relative concentration of the substrates and products of the reactions involved.[5]:721
Alanine World Hypothesis
Alanine is one of the twenty canonical α-amino acids used as building blocks (monomers) for the ribosome-mediated biosynthesis of proteins. Alanine is believed to be one of the earliest amino acids to be included in the genetic code standard repertoire.[16][17][18][7] On the basis of this fact the "Alanine World" hypothesis was proposed.[19] This hypothesis explains the evolutionary choice of amino acids in the repertoire of the genetic code from a chemical point of view. In this model the selection of monomers (i.e. amino acids) for ribosomal protein synthesis is rather limited to those Alanine derivatives that are suitable for building α-helix or β-sheet secondary structural elements. Dominant secondary structures in life as we know it are α-helices and β-sheets and most canonical amino acids can be regarded as chemical derivatives of Alanine. Therefore, most canonical amino acids in proteins can be exchanged with Ala by point mutations while the secondary structure remains intact. The fact that Ala mimics the secondary structure preferences of the majority of the encoded amino acids is practically exploited in alanine scanning mutagenesis. In addition, classical X-ray crystallography often employs the polyalanine-backbone model[20] to determine three-dimensional structures of proteins using molecular replacement - a model-based phasing method.
Physiological function
Glucose–alanine cycle
In mammals, alanine plays a key role in glucose–alanine cycle between tissues and liver. In muscle and other tissues that degrade amino acids for fuel, amino groups are collected in the form of glutamate by transamination. Glutamate can then transfer its amino group to pyruvate, a product of muscle glycolysis, through the action of alanine aminotransferase, forming alanine and α-ketoglutarate. The alanine enters the bloodstream, and is transported to the liver. The alanine aminotransferase reaction takes place in reverse in the liver, where the regenerated pyruvate is used in gluconeogenesis, forming glucose which returns to the muscles through the circulation system. Glutamate in the liver enters mitochondria and is broken down by glutamate dehydrogenase into α-ketoglutarate and ammonium, which in turn participates in the urea cycle to form urea which is excreted through the kidneys.[21]
The glucose–alanine cycle enables pyruvate and glutamate to be removed from muscle and safely transported to the liver. Once there, pyruvate is used to regenerate glucose, after which the glucose returns to muscle to be metabolized for energy: this moves the energetic burden of gluconeogenesis to the liver instead of the muscle, and all available ATP in the muscle can be devoted to muscle contraction.[21] It is a catabolic pathway, and relies upon protein breakdown in the muscle tissue. Whether and to what extent it occurs in non-mammals is unclear.[22][23]
Link to diabetes
Alterations in the alanine cycle that increase the levels of serum alanine aminotransferase (ALT) are linked to the development of type II diabetes.[24]
Chemical properties
Alanine is useful in loss of function experiments with respect to phosphorylation. Some techniques involve creating a library of genes, each of which has a point mutation at a different position in the area of interest, sometimes even every position in the whole gene: this is called "scanning mutagenesis". The simplest method, and the first to have been used, is so-called alanine scanning, where every position in turn is mutated to alanine.[25]
Hydrogenation of alanine give the amino alcohol alaninol, which is a useful chiral building block.
Free radical
The deamination of an alanine molecule produces the free radical CH3C•HCO2−. Deamination can be induced in solid or aqueous alanine by radiation that causes homolytic cleavage of the carbon–nitrogen bond.[26]
This property of alanine is used in dosimetric measurements in radiotherapy. When normal alanine is irradiated, the radiation causes certain alanine molecules to become free radicals, and, as these radicals are stable, the free radical content can later be measured by electron paramagnetic resonance in order to find out how much radiation the alanine was exposed to.[27] This is considered to be a biologically relevant measure of the amount of radiation damage that living tissue would suffer under the same radiation exposure.[27] Radiotherapy treatment plans can be delivered in test mode to alanine pellets, which can then be measured to check that the intended pattern of radiation dose is correctly delivered by the treatment system.
References
- "L-alanine_msds".
- Haynes, William M., ed. (2016). CRC Handbook of Chemistry and Physics (97th ed.). CRC Press. pp. 5–88. ISBN 978-1498754286.
- "Nomenclature and Symbolism for Amino Acids and Peptides". IUPAC-IUB Joint Commission on Biochemical Nomenclature. 1983. Archived from the original on 9 October 2008. Retrieved 5 March 2018.
- Doolittle, Russell F. (1989). "Redundancies in Protein Sequences". In Fasman, Gerald D. (ed.). Prediction of Protein Structures and the Principles of Protein Conformation. New York: Plenum. pp. 599–623. ISBN 0-306-43131-9.
- Mathews, Christopher K.; Van Holde, Kensal Edward; Ahern, Kevin G. (2000). Biochemistry (3rd ed.). San Francisco, CA: Benjamin/Cummings Publishing. ISBN 0805330666. OCLC 42290721.
- Yoshikawa, Naoko; Sarower, Mohammed G.; Abe, Hiroki (2016). "Alanine Racemase and D-Amino Acid Oxidase in Aquatic Animals". In Yoshimura, Tohru; Nishikawa, Toru; Homma, Hiroshi (eds.). D-Amino Acids: Physiology, Metabolism, and Application. Springer Japan. pp. 269–282. ISBN 9784431560777.
- Ntountoumi, Chrysa; Vlastaridis, Panayotis; Mossialos, Dimitris; Stathopoulos, Constantinos; Iliopoulos, Ioannis; Promponas, Vasilios; Oliver, Stephen G; Amoutzias, Grigoris D (2019-11-04). "Low complexity regions in the proteins of prokaryotes perform important functional roles and are highly conserved". Nucleic Acids Research. 47 (19): 9998–10009. doi:10.1093/nar/gkz730. ISSN 0305-1048. PMC 6821194. PMID 31504783.
- Strecker, Adolph (1850). "Ueber die künstliche Bildung der Milchsäure und einen neuen, dem Glycocoll homologen" [On the artificial formation of lactic acid and a new substance homologous to glycine]. Annalen der Chemie und Pharmacie (in German). 75 (1): 27–45. doi:10.1002/jlac.18500750103. Strecker names alanine on p. 30.
- Strecker, Adolph (1854). "Ueber einen neuen aus Aldehyd – Ammoniak und Blausäure entstehenden Körper" [On a new substance arising from acetaldehyde–ammonia [i.e., 1-aminoethanol] and hydrocyanic acid]. Annalen der Chemie und Pharmacie (in German). 91 (3): 349–351. doi:10.1002/jlac.18540910309.
- "Alanine". AminoAcidsGuide.com. 10 June 2018. Retrieved 14 April 2019.
- "alanine". Oxford Dictionaries. Retrieved 2015-12-06.
- Textbook of Biotechnology. McGraw-Hill Education (I). 2012. ISBN 9780071070072.
- Karlheinz Drauz; Ian Grayson; Axel Kleemann; Hans-Peter Krimmer; Wolfgang Leuchtenberger; Christoph Weckbecker (2006). Ullmann's Encyclopedia of Industrial Chemistry. Weinheim: Wiley-VCH. doi:10.1002/14356007.a02_057.pub2.
- Kendall, E. C.; McKenzie, B. F. (1929). "dl-Alanine". Organic Syntheses. 9: 4. doi:10.15227/orgsyn.009.0004.; Collective Volume, 1, p. 21.
- Tobie, Walter C.; Ayres, Gilbert B. (1941). "dl-Alanine". Organic Syntheses. doi:10.15227/orgsyn.009.0004.; Collective Volume, 1, p. 21
- Trifonov, E.N (December 2000). "Consensus temporal order of amino acids and evolution of the triplet code". Gene. 261 (1): 139–151. doi:10.1016/S0378-1119(00)00476-5.
- Higgs, Paul G.; Pudritz, Ralph E. (June 2009). "A Thermodynamic Basis for Prebiotic Amino Acid Synthesis and the Nature of the First Genetic Code". Astrobiology. 9 (5): 483–490. arXiv:0904.0402. doi:10.1089/ast.2008.0280. ISSN 1531-1074.
- Kubyshkin, Vladimir; Budisa, Nediljko (24 September 2019). "The Alanine World Model for the Development of the Amino Acid Repertoire in Protein Biosynthesis". Int. J. Mol. Sci. 20 (21): 5507. doi:10.3390/ijms20215507. PMC 6862034. PMID 31694194.
- Kubyshkin, Vladimir; Budisa, Nediljko (3 July 2019). "Anticipating alien cells with alternative genetic codes: away from the alanine world!". Curr. Op. Biotechnol. 60: 242–249. doi:10.1016/j.copbio.2019.05.006. PMID 31279217.
- Karmali, A. M.; Blundell, T. L.; Furnham, N. (2009). "Model-building strategies for low-resolution X-ray crystallographic data". Acta Crystallogr. 65 (2): 121–127. doi:10.1107/S0907444908040006. PMC 2631632. PMID 19171966.
- Nelson, David L.; Cox, Michael M. (2005). Principles of Biochemistry (4th ed.). New York: W. H. Freeman. pp. 684–85. ISBN 0-7167-4339-6..
- Fish Physiology: Nitrogen Excretion. Academic Press. 2001-09-07. p. 23. ISBN 9780080497518.
- Walsh, Patrick J.; Wright, Patricia A. (1995-08-31). Nitrogen Metabolism and Excretion. CRC Press. ISBN 9780849384110.
- Sattar, Naveed; Scherbakova, Olga; Ford, Ian; O'Reilly, Denis St. J.; Stanley, Adrian; Forrest, Ewan; MacFarlane, Peter W.; Packard, Chris J.; Cobbe, Stuart M.; Shepherd, James (2004). "Elevated Alanine Aminotransferase Predicts New-Onset Type 2 Diabetes Independently of Classical Risk Factors, Metabolic Syndrome, and C-Reactive Protein in the West of Scotland Coronary Prevention Study". Diabetes. 53 (11): 2855–2860. doi:10.2337/diabetes.53.11.2855. PMID 15504965.
- Park, Sheldon J.; Cochran, Jennifer R. (2009-09-25). Protein Engineering and Design. CRC Press. ISBN 9781420076592.
- Zagórski, Z. P.; Sehested, K. (1998). "Transients and Stable Radical from the Deamination of α-Alanine". J. Radioanal. Nucl. Chem. 232 (1–2): 139–41. doi:10.1007/BF02383729. S2CID 97855573..
- Pedro, Andreo; Burns, David T.; Nahum, Alan E.; Seuntjens, Jan P.; Attix, Frank H. (2017). "Alanine Dosimetry". Fundamentals of Ionizing Radiation Dosimetry (2nd ed.). Weinheim, Germany: Wiley-VCH. pp. 547–556. ISBN 9783527808236. OCLC 990023546.
External links
- Alanine MS spectrum
- 3-Hydroxyphosphinylalanine[1]
- Pubchem. "Alanine, 3-(hydroxyphosphinyl)-". pubchem.ncbi.nlm.nih.gov. Retrieved 20 September 2018.