DPANN
DPANN is a superphylum of Archaea first proposed in 2013. Many members show novel signs of horizontal gene transfer from other domains of life.[2] They are known as nanoarchaea or ultra-small archaea due to their smaller size (nanometric) compared to other archaea.
| DPANN | |
|---|---|
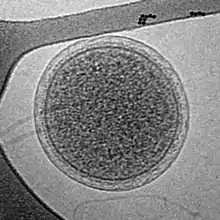 | |
| Parvarchaeum acidiphilum | |
| Scientific classification | |
| Domain: | |
| Superphylum: | DPANN group |
| Phyla[1] | |
| |
DPANN is an acronym formed by the initials of the first five groups discovered, Diapherotrites, Parvarchaeota, Aenigmarchaeota, Nanoarchaeota and Nanohaloarchaeota. Later Woesearchaeota and Pacearchaeota were discovered and proposed within the DPANN superphylum.[3] In 2017, another phylum Altiarchaeota was placed into this superphylum.[4]
The DPANN groups together different phyla with a variety of environmental distribution and metabolism, ranging from symbiotic and thermophilic forms such as Nanoarchaeota, acidophiles like Parvarchaeota and non-extremophiles like Aenigmarchaeota and Diapherotrites. DPANN was also detected in nitrate-rich groundwater, on the water surface but not below, indicating that these taxa are still quite difficult to locate.[5]
Characteristcs
They are characterized by being small in size compared to other archaea (nanometric size) and in keeping with their small genome, they have limited but sufficient catabolic capacities to lead a free life, although many are episymbionts that depend on a symbiotic or parasitic association with other organisms. Many of its characteristics are similar or analogous to those of ultra-small bacteria (CPR group).[3]
Limited metabolic capacities are a product of the small genome and are reflected in the fact that many lack central biosynthetic pathways for nucleotides, aminoacids, and lipids; hence most DPANN archaea, such as ARMAN archaea, which rely on other microbes to meet their biological requirements. But those that have the potential to live freely are fermentative and aerobic heterotrophs.[3]
They are mostly anaerobic that cannot be cultivated. They live in extreme environments such as thermophilic, hyperacidophilic, hyperhalophilic or metal-resistant; or also in the temperate environment of marine and lake sediments. They are rarely found on the ground or in the open ocean.[3]
Classification
- Diapherotrites. Found by phylogenetic analysis of the genomes recovered from the groundwater filtration of a gold mine abandoned in the USA.[6][7]
- Parvarchaeota and Micrarchaeota. Discovered in 2006 in acidic mine drainage from a US mine.[8][9][10] They are of very small size and provisionally called ARMAN (Archaeal Richmond Mine Acidophilic Nanoorganisms).
- Woesearchaeota and Pacearchaeota. They have been identified both in sediments and in surface waters of aquifers and lakes, abounding especially in saline conditions.[3][11]
- Aenigmarchaeota. Found in wastewater from mines and in sediments from hot springs.[12]
- Nanohaloarchaeota. Distributed in environments with high salinity.[13]
- Nanoarchaeota. They were the first discovered (in 2002) in a hydrothermal source next to the coast of Iceland. They live as symbionts of other archaea.[14][15]
Phylogeny
DPANN may be the first divergent clade of archaea according to some phylogenetic analyzes. Recent phylogenetic analyzes have found the following phylogeny between phyla.[3][16][17]
| Archaea |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Other phylogenetic analyzes have suggested that DPANN could belong to Euryarchaeota or that it may even be polyphyletic occupying different positions within Euryarchaeota. It is also debated whether the phylum Altiarchaeota should be classified in DPANN or Euryarchaeota.[17][18] An alternative location for DPANN in the phylogenetic tree is as follows.[19][20] The groups marked in quotes are lineages assigned to DPANN, but phylogenetically separated from the rest.
| Archaea |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
References
- Castelle CJ, Banfield JF (2018). "Major New Microbial Groups Expand Diversity and Alter our Understanding of the Tree of Life". Cell. 172 (6): 1181–1197. doi:10.1016/j.cell.2018.02.016. PMID 29522741.
- Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng JF, Darling A, Malfatti S, Swan BK, Gies EA, Dodsworth JA, Hedlund BP, Tsiamis G, Sievert SM, Liu WT, Eisen JA, Hallam SJ, Kyrpides NC, Stepanauskas R, Rubin EM, Hugenholtz P, Woyke T (July 2013). "Insights into the phylogeny and coding potential of microbial dark matter" (PDF). Nature. 499 (7459): 431–437. doi:10.1038/nature12352. PMID 23851394.
- Castelle CJ, Wrighton KC, Thomas BC, Hug LA, Brown CT, Wilkins MJ, Frischkorn KR, Tringe SG, Singh A, Markillie LM, Taylor RC, Williams KH, Banfield JF (March 2015). "Genomic expansion of domain archaea highlights roles for organisms from new phyla in anaerobic carbon cycling". Current Biology. 25 (6): 690–701. doi:10.1016/j.cub.2015.01.014. PMID 25702576.
- Spang A, Caceres EF, Ettema TJ (August 2017). "Genomic exploration of the diversity, ecology, and evolution of the archaeal domain of life". Science. 357 (6351): eaaf3883. doi:10.1126/science.aaf3883. PMID 28798101.
- Ludington WB, Seher TD, Applegate O, Li X, Kliegman JI, Langelier C, Atwill ER, Harter T, DeRisi JL (2017-04-06). "Assessing biosynthetic potential of agricultural groundwater through metagenomic sequencing: A diverse anammox community dominates nitrate-rich groundwater". PLOS One. 12 (4): e0174930. doi:10.1371/journal.pone.0174930. PMC 5383146. PMID 28384184.
- Genomes Online Database
- Comolli LR, Baker BJ, Downing KH, Siegerist CE, Banfield JF (February 2009). "Three-dimensional analysis of the structure and ecology of a novel, ultra-small archaeon". The ISME Journal. 3 (2): 159–167. doi:10.1038/ismej.2008.99. PMID 18946497.
- Baker BJ, Tyson GW, Webb RI, Flanagan J, Hugenholtz P, Allen EE, Banfield JF (December 2006). "Lineages of acidophilic archaea revealed by community genomic analysis". Science. 314 (5807): 1933–1935. doi:10.1126/science.1132690. PMID 17185602.
- Murakami S, Fujishima K, Tomita M, Kanai A (February 2012). "Metatranscriptomic analysis of microbes in an Oceanfront deep-subsurface hot spring reveals novel small RNAs and type-specific tRNA degradation". Applied and Environmental Microbiology. 78 (4): 1015–1022. doi:10.1128/AEM.06811-11. PMC 3272989. PMID 22156430.
- Baker BJ, Comolli LR, Dick GJ, Hauser LJ, Hyatt D, Dill BD, Land ML, Verberkmoes NC, Hettich RL, Banfield JF (May 2010). "Enigmatic, ultrasmall, uncultivated Archaea". Proceedings of the National Academy of Sciences of the United States of America. 107 (19): 8806–8811. doi:10.1073/pnas.0914470107. PMC 2889320. PMID 20421484.
- Ortiz-Alvarez R, Casamayor EO (April 2016). "High occurrence of Pacearchaeota and Woesearchaeota (Archaea superphylum DPANN) in the surface waters of oligotrophic high-altitude lakes". Environmental Microbiology Reports. 8 (2): 210–217. doi:10.1111/1758-2229.12370. PMID 26711582.
- Takai K, Moser DP, DeFlaun M, Onstott TC, Fredrickson JK (December 2001). "Archaeal diversity in waters from deep South African gold mines". Applied and Environmental Microbiology. 67 (12): 5750–5760. doi:10.1128/AEM.67.21.5750-5760.2001. PMC 93369. PMID 11722932.
- Narasingarao P, Podell S, Ugalde JA, Brochier-Armanet C, Emerson JB, Brocks JJ, Heidelberg KB, Banfield JF, Allen EE (January 2012). "De novo metagenomic assembly reveals abundant novel major lineage of Archaea in hypersaline microbial communities". The ISME Journal. 6 (1): 81–93. doi:10.1038/ismej.2011.78. PMC 3246234. PMID 21716304.
- Waters E, Hohn MJ, Ahel I, Graham DE, Adams MD, Barnstead M, Beeson KY, Bibbs L, Bolanos R, Keller M, Kretz K, Lin X, Mathur E, Ni J, Podar M, Richardson T, Sutton GG, Simon M, Soll D, Stetter KO, Short JM, Noordewier M (October 2003). "The genome of Nanoarchaeum equitans: insights into early archaeal evolution and derived parasitism". Proceedings of the National Academy of Sciences of the United States of America. 100 (22): 12984–12988. doi:10.1073/pnas.1735403100. PMC 240731. PMID 14566062.
- Podar M, Makarova KS, Graham DE, Wolf YI, Koonin EV, Reysenbach AL (December 2013). "Insights into archaeal evolution and symbiosis from the genomes of a nanoarchaeon and its inferred crenarchaeal host from Obsidian Pool, Yellowstone National Park". Biology Direct. 8 (1): 9. doi:10.1186/1745-6150-8-9. PMC 3655853. PMID 23607440.
- Williams TA, Szöllősi GJ, Spang A, Foster PG, Heaps SE, Boussau B, et al. (June 2017). "Integrative modeling of gene and genome evolution roots the archaeal tree of life". Proceedings of the National Academy of Sciences of the United States of America. 114 (23): E4602–E4611. doi:10.1073/pnas.1618463114. PMC 5468678. PMID 28533395.
- Dombrowski N, Williams TA, Sun J, Woodcroft BJ, Lee JH, Minh BQ, et al. (August 2020). "Undinarchaeota illuminate DPANN phylogeny and the impact of gene transfer on archaeal evolution". Nature Communications. 11 (1): 3939. doi:10.1038/s41467-020-17408-w. PMC 7414124. PMID 32770105.
- Nina Dombrowski, Jun-Hoe Lee, Tom A Williams, Pierre Offre, Anja Spang (2019). Genomic diversity, lifestyles and evolutionary origins of DPANN archaea. Nature.
- Jordan T. Bird, Brett J. Baker, Alexander J. Probst, Mircea Podar, Karen G. Lloyd (2017). Culture Independent Genomic Comparisons Reveal Environmental Adaptations for Altiarchaeales. Frontiers.
- Thomas Cavalier-Smith & Ema E-Yung Chao (2020). Multidomain ribosomal protein trees and the planctobacterial origin of neomura (eukaryotes, archaebacteria). Linkspringer.
External links
 Data related to DPANN group at Wikispecies
Data related to DPANN group at Wikispecies
